Abstract
We developed a multiplex PCR-based methodology for nasopharyngeal samples maintained in egg thioglycolate antibiotic and skim milk-tryptone-glucose-glycerol media to identify and serotype the most important serotypes of Streptococcus pneumoniae that cause invasive disease in children. This technique can be used to study the epidemiology of pneumococcal colonization and the effect of conjugate vaccines.
Multiple serotypes of Streptococcus pneumoniae can simultaneously colonize the nasopharynx of children (15). Identification of these serotypes is important for surveillance programs and for evaluation of the effect of vaccination on nasopharyngeal carriage (8). Recent evidence shows that a decrease in carriage of vaccine serotypes and a significant increase of nonvaccine serotypes occurs in immunized children, probably due to replacement of serotypes or unmasking of minority populations of S. pneumoniae present in the nasopharynx. This could cause important changes in the epidemiology of pneumococcal invasive diseases (2, 6).
The purpose of this study was to develop a molecular methodology to identify S. pneumoniae and the simultaneous carriage of serotypes from a single nasopharyngeal sample. We used PCR to screen the autolysin gene (lytA) (11, 12) and a multiplex PCR to amplify specific regions of the capsular genes of serogroups 6 and 18 and serotypes 1, 3, 4, 14, 19A, 19F, and 23F (1, 9). These capsular types are responsible for the majority of invasive diseases in children less than 5 years old (5, 15) and are frequently isolated from nasopharynx (10, 16, 17, 19, 21), and the greater part of these are included in the heptavalent conjugate vaccine (8, 15).
PCR assays to amplify lytA were performed as described previously, using the outer primers only (11). Serogroup and serotype primer sets used in the multiplex PCR have been previously published (1, 9) and were distributed in two mixtures (Fig. 1). Mixture 1 had primers for serogroups 1 (103 bp), 3 (152 bp), 6 (220 bp), and 18 (354 bp) and serotype 19A (478 bp); mixture 2 had primers for serotypes 19F (130 bp) and 23F (177 bp) and serogroups 14 (220 bp) and 4 (430 bp). PCRs were performed in 25-μl reaction volumes that contained 1× PCR buffer, 2.4 mM MgCl2, 240 nM of each deoxynucleoside triphosphate, 2 U of Taq polymerase (Invitrogen), different concentrations of each primer (35 nM to 178 nM), and 2 μl of DNA suspension. The multiplex PCR was performed on a PTC-225 (MJ Research) as follows: 95°C for 5 min for one cycle followed by 35 cycles at 95°C for 30 s, 61°C for 45 s, and 72°C for 1 min, with the last cycle at 72°C for 5 min. The amplified products were analyzed by 1.8% agarose gel electrophoresis with 0.2 μg/ml ethidium bromide, and serotypes were identified by visualization of the equivalent band to the molecular weight of the corresponding gene.
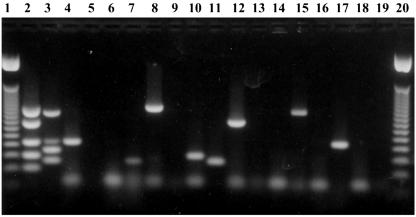
FIG. 1.
Multiplex PCR for serotyping Streptococcus pneumoniae from nasopharyngeal samples. Lanes 1 and 20, molecular weight of 50 bp; lane 2, mixture 1; lane 3, mixture 2; lanes 4 and 5, sample 10 (ETA); lanes 6 and 7, sample 13 (ETA); lanes 8 and 9, sample 5 (STGG); lanes 10 and 11, sample 6 (STGG); lanes 12 and 13, sample 8 (STGG); lanes 14 and 15, sample 9 (STGG); lanes 16 and 17, sample 10 (STGG); and lanes 18 and 19, negative controls.
Specificity tests were performed with 33 different serotypes of S. pneumoniae (1, 2, 3, 4, 5, 6A, 6B, 7F, 8, 9N, 9V, 10, 11, 13, 14, 15A, 15B, 15C, 16F, 18A, 18B, 18C, 19A, 19B, 19F, 22, 23A, 23B, 23C, 23F, 34, 35, and 35B), S. intermedius, S. sanguis, S. salivarius, S. canis, S. pyogenes, S. bovis, S. agalactiae, S. mitis, S. equis, Staphylococcus aureus, S. epidermidis, Enterococcus faecalis, Corynebacterium diphteriae, Haemophilus influenzae type b, Moraxella catarrhalis, Neisseria lactamica, and N. meningitidis.
The detection limit of the PCR for the autolysin gene was 500 fg of DNA or about 400 genome equivalents, and that of the multiplex PCR fell in a range between 39 (serotype 19F) and 246 CFU (serogroup 18) or about 49 and 308 fg, respectively. The specificity tests showed no cross-reaction with any of the PCR assays.
To evaluate the methodology with nasopharyngeal specimens, calcium alginate swabs were obtained from 19 children between 10 and 24 months of age that attend a day care center and had previously given informed consent. Samples were immediately plated onto 5% sheep blood agar and 5.0 μg/ml gentamicin and were processed by standardized methodology (15). Swabs were inoculated into egg thioglycolate antibiotic (ETA) transport and storage media (4) and were maintained at room temperature for several days before PCRs.
The swab used for PCR was placed in 100 μl of physiological saline, 20 μl of this suspension was stored at −70°C in skim milk-tryptone-glucose-glycerol (STGG) transport media (14), and 10 μl was enriched by inoculating into 1.2 ml of Todd-Hewitt broth supplemented with 22 μg/ml glutamine and 5.0 μg/ml gentamicin and incubated for 4 h in a 37°C water bath (13). The cells were recovered by centrifugation (10,000 rpm for 10 min) and resuspended in 100 μl of physiological saline, and DNA was extracted by boiling for 10 min.
Additionally, we used 11 nasopharyngeal samples obtained from children less than 5 years old, with informed consent, in a study carried out in a day care center in 1999 and stored at −70°C in STGG medium (3). From them, 50 μl was plated onto 5% sheep blood agar and 5.0 μg/ml gentamicin for recovery and serotype, and 50 μl was used for DNA extraction according to the previously described methodology.
The multiplex PCR identified the S. pneumoniae serotypes present in the nasopharyngeal samples maintained either in ETA or STGG medium (Fig. 1). In 14 out of 19 nasopharyngeal samples stored in ETA, S. pneumoniae was recovered by culture and identified by amplification of the lytA gene. The PCR multiplex correctly identified 10 of 11 serotypes and serogroups that could be detected and agreed with the results obtained by the Quellung reaction (Table 1). In sample 13, the multiplex PCR identified a second serotype (19F) not detected by culture, which was confirmed from the sample conserved in STGG medium.
TABLE 1.
Results of culture methodology, Quellung reaction, and PCR assays for detection and serotyping of S. pneumoniae from nasopharyngeal samples stored in ETA transport mediaa
| Sample no. | Culture
|
PCR
|
||
|---|---|---|---|---|
| Recovery of S. pneumoniae | Quellung reaction | lytA | Serotype | |
| 1 | + | 6A | + | 6 |
| 2 | SGV | − | ||
| 3 | + | 6A | + | 6 |
| 4 | + | 6A | + | 6 |
| 5 | + | 6A | + | 6 |
| 6 | NG | − | ||
| 7 | + | 6A | + | 6 |
| 8 | SGV | − | ||
| 9 | + | 6A, NT | + | 6 |
| 10 | + | 6A | + | 6 |
| 11 | + | 6A, NT | + | 6 |
| 12 | + | 15B | + | |
| 13 | + | 15C | + | 19F |
| 14 | + | 6B | + | 6 |
| 15 | + | 6B | + | 6 |
| 16 | NG | − | ||
| 17 | + | 19F, NT | + | |
| 18 | + | 35, NT | + | |
| 19 | SGV | − | ||
NG, no growth; SGV, S. viridans group; NT, nontypeable S. pneumoniae.
The discordant result corresponded to sample 17, in which the majority of colonies were nontypeable S. pneumoniae and only a single colony of 19F was recovered from blood agar. The failure of the multiplex PCR to detect serotype 19F probably was due to growth of nasopharyngeal contaminants during the enrichment step that could have inhibited growth of the streptococcus population (20). It is also possible that there might have been an error during processing of the sample.
All samples stored in STGG were amplified with the PCR for lytA, including sample 11, in which S. pneumoniae was not recovered in culture. In nine samples the results of the PCR multiplex and Quellung reaction agreed. However, PCR identified a second serotype in sample 6, but in sample 5 differences in the determination of specific serotype were observed (Table 2). Originally, samples 6 and 11 had low colony counts (3), and under these conditions the recovery of pneumococci after storage of material in STGG medium at 70°C was reduced (14). These results suggest that minority populations in nasopharyngeal samples can be detected by PCR compared with culture (18).
TABLE 2.
Results of culture methodology, Quellung reaction, and PCR assays for detection and serotyping of S. pneumoniae from nasopharyngeal samples stored in STGG mediuma
| Sample no. | Culture
|
Serotype by PCR | |
|---|---|---|---|
| Recovery of S. pneumoniae | Quellung reaction | ||
| 1 | + | 3 | 3 |
| 2 | + | 3 | 3 |
| 3 | + | 4 | 4 |
| 4 | + | 3 | 3 |
| 5 | + | 19F | 19A |
| 6 | + | 19F | 3, 19F |
| 7 | + | 3 | 3 |
| 8 | + | 18 | 18 |
| 9 | + | 4 | 4 |
| 10 | + | 14 | 14 |
| 11 | NG | 19A | |
NG, no growth; all samples were positive for lytA by PCR.
In sample 5, PCR detected serotype 19A, and by Quellung serotype 19F was identified. The PCR result was verified from the colony isolated by Quellung and multiplex PCR using only the primers for serotypes 19A and 19F. Moreover, the primers for serotypes 19A and 19F were evaluated with isolates of both serotypes obtained in our laboratory, two isolates from the National Center for Streptococcus of Canada, and the clones S. Africa19A-13 and Taiwan19F-14 without observing cross-reaction. We do not have an explanation for these results yet.
In summary, we have developed a methodology that permits the detection and identification of some relevant serotypes and serogroups from nasopharyngeal samples maintained in STGG or ETA. This technique has the advantage of being able to detect a second serotype of pneumococci in nasopharyngeal samples, in contrast with the culture method that has limited sensitivity for detecting a second serotype present in smaller proportions (7, 15). Moreover, the multiplex PCR assay can readily be extended to include serotype 9V and other capsule loci as sequences become available and according to the regional epidemiologic distribution of the serotypes. However, the technique should be evaluated with a higher number of nasopharyngeal samples to be used in epidemiological studies.
REFERENCES
- 1.Brito, D., M. Ramírez, and H. Lencastre. 2003. Serotyping Streptococcus pneumoniae by multiplex PCR. J. Clin. Microbiol. 41:2378-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byngton, C. L., M. H. Samore, G. J. Stoddard, S. Barlow, J. Dalay, K. Korgenski, S. Firth, D. Glover, J. Jensen, E. O. Masson, C. K. Shutt, and A. Pavia. 2005. Temporal trends of invasive disease due to Streptococcus pneumoniae among children in the intermountain west: emergence of nonvaccine serogroups. Clin. Infect. Dis. 41:21-29. [DOI] [PubMed] [Google Scholar]
- 3.Fonseca, N., M. C. Vela, and E. Castañeda. 2000. Evaluación de un medio para la conservación de Streptococcus pneumoniae a partir de muestras nasofaríngeas. Biomédica 20:261-264. [Google Scholar]
- 4.Gray, B. M. 2002. Egg-based media for delayed processing of nasopharyngeal swabs in colonization studies of Streptococcus pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 21:666-670. [DOI] [PubMed] [Google Scholar]
- 5.Hausdorff, W. P., J. Bryant, P. R. Paradiso, and G. R. Siber. 2000. The contribution of specific pneumococcal serogroups to different disease manifestations: implications for conjugate vaccine formulation and use, part I. Clin. Infect. Dis. 30:100-121. [DOI] [PubMed] [Google Scholar]
- 6.Huang, S. S., R. Platt, S. L. Rifas-Shiman, S. I. Pelton, D. Goldmann, and J. A. Finkeslstein. 2005. Post-PCV7 changes in colonization pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics 116:e408-e413. [DOI] [PubMed] [Google Scholar]
- 7.Huebner, R. E., R. Dagan, N. Porath, A. D. Wasas, and K. P. Klugman. 2000. Lack of utility of serotyping multiple colonies for detection of simultaneous nasopharyngeal carriage of different pneumococcal serotypes. Pediatr. Infect. Dis. J. 19:1017-1019. [DOI] [PubMed] [Google Scholar]
- 8.Klugman, K. P. 2001. Efficacy of pneumococcal conjugate vaccines and their effect on carriage and antimicrobial resistance. Lancet Infect. Dis. 1:85-91. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence, E., D. Griffiths, S. Martin, R. George, and L. Hall. 2003. Evaluation of semiautomated multiplex PCR assay for determination of Streptococcus pneumoniae serotypes and serogroups. J. Clin. Microbiol. 41:601-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malfroot, A., J. Verhaegen, J. M. Dubru, E. Van Kerschaver, and S. Leyman. 2004. A cross-sectional survey of the prevalence of Streptococcus pneumoniae nasopharyngeal carriage in Belgian infants attending day care centres. Clin. Microbiol. Infect. 10:797-803. [DOI] [PubMed] [Google Scholar]
- 11.Messmer, T. O., C. G. Whitney, and B. S. Fields. 1997. Use of polymerase chain reaction to identify pneumococcal infection associated with hemorrhage and shock in two previously healthy young children. Clin. Chem. 43:930-935. [PubMed] [Google Scholar]
- 12.Messmer, T. O., J. S. Sampson, A. Stinson, B. Wong, G. M. Carlone, and R. Facklam. 2004. Comparison of four polymerase chain reaction assays for specificity in the identification of Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 49:249-254. [DOI] [PubMed] [Google Scholar]
- 13.Morrison, K., D. Lake, J. Crook, G. Carlone, E. Ades, R. Facklam, and J. Sampson. 2000. Confirmation of psaA in all 90 serotypes of Streptococcus pneumoniae by PCR and potential of this assay for identification and diagnosis. J. Clin. Microbiol. 38:434-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Brien, K., M. A. Bronsdon, R. Dagan, P. Yagupsky, J. Janco, J. Elliott, C. G. Whitney, Y. Yang, L. E. Robinson, B. Schwartz, and G. M. Carlone. 2001. Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J. Clin. Microbiol. 39:1021-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Brien, K., H. Nohynek, and the WHO Pneumococcal Vaccine Trials Carriage Working Group. 2003. Report from a WHO Working Group: standard method for detecting upper respiratory carriage of Streptococcus pneumoniae. Pediatr. Infect. Dis. J. 22:133-140. [DOI] [PubMed] [Google Scholar]
- 16.Pons, J. L., M. N. Mandement, E. Martin, C. Lemort, M. Nouvellon, E. Mallet, and J. F. Lemeland. 1996. Clonal and temporal patterns of nasopharyngeal penicillin-susceptible and penicillin-resistant Streptococcus pneumoniae strains in children attending a day care center. J. Clin. Microbiol. 34:3218-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sá-Leāo, R., A. Tomasz, I. S. Sanches, S. Nunes, C. R. Alves, A. B. Avô, J. Saldaña, K. Kristinsson, and H. de Lencastre. 2000. Genetic diversity and clonal patterns among antibiotic-susceptible and -resistant Streptococcus pneumoniae colonizing children: day care centers as autonomous epidemiological units. J. Clin. Microbiol. 38:4137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saukkoriipi, A., K. Leskela, E. Herva, and M. Leinonen. 2004. Streptococcus pneumoniae in nasopharyngeal secretions of healthy children: comparison of real-time PCR and culture from STGG-transport medium. Mol. Cell Prob. 18:147-153. [DOI] [PubMed] [Google Scholar]
- 19.Syrjanen, R. K., T. M. Kilpi, T. H. Kaijalainen, E. E. Herva, and A. K. Takala. 2001. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J. Infect. Dis. 184:451-459. [DOI] [PubMed] [Google Scholar]
- 20.Tano, K., E. Grahn Hakansson, S. E. Holm, and S. Hellstrom. 2000. Inhibition of OM pathogens by alpha-hemolytic streptococci from healthy children, children with SOM and children with rAOM. Int. J. Pedriatr. Otorhinolaryngol. 56:185-190. [DOI] [PubMed] [Google Scholar]
- 21.Yagupsky, P., N. Porat, D. Fraser, F. Prajgrod, M. Merires, L. McGee, K. P. Klugman, and R. Dagan. 1996. Acquisition, carriage, and transmission of pneumococci with decreased antibiotic susceptibility in young children attending a day care facility in southern Israel. J. Infect. Dis. 157:1003-1012. [DOI] [PubMed] [Google Scholar]