Fig. 4. Delineation of a binding site within the region between −84 and −34 that binds nuclear factors from keratinocytes.
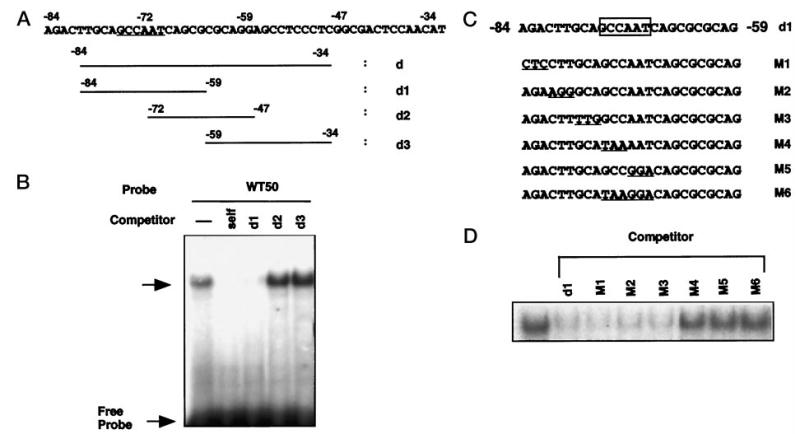
A, the DNA sequence between −84 and −34 (oligomer d) and oligomers used in the competitive gel shift assay (d1–d3); the CCAAT box is underlined. B, the d oligomer (−84/−34) was labeled and used as a probe in gel shift assay with nuclear extracts of keratinocytes (3 μg). In competition assays, the nuclear extracts were mixed with each competing oligonucleotide (100-fold molar excess) prior to addition of the 32P-labeled probe. The data presented in this figure are representative of electromobility shift assays that were performed at least three separate times with the same results. C, the DNA sequence for oligonucleotide d1 is shown, along with the location of the 3-bp mutations. The putative CCAAT motif is boxed. The serial 3-bp mutated sequences are underlined in each mutated oligomer (M1–M6). D, competition binding assay with 32P-labeled d1 oligomer used as a probe and competitor oligomers at 100-fold molar excess (d1 and M1–M6). Identical results were observed with multiple nuclear extract preparations.
