Abstract
We describe a novel quantitative real-time (Q)-PCR assay for Listeria monocytogenes based on the coamplification of a target hly gene fragment and an internal amplification control (IAC). The IAC is a chimeric double-stranded DNA containing a fragment of the rapeseed BnACCg8 gene flanked by the hly-specific target sequences. This IAC is detected using a second TaqMan probe labeled with a different fluorophore, enabling the simultaneous monitoring of the hly and IAC signals. The hly-IAC assay had a specificity and sensitivity of 100%, as assessed using 49 L. monocytogenes isolates of different serotypes and 96 strains of nontarget bacteria, including 51 Listeria isolates. The detection and quantification limits were 8 and 30 genome equivalents, and the coefficients for PCR linearity (R2) and efficiency (E) were 0.997 and 0.80, respectively. We tested the performance of the hly-IAC Q-PCR assay using various broth media and food matrices. Fraser and half-Fraser media, raw pork, and raw or cold-smoked salmon were strongly PCR-inhibitory. This Q-PCR assay for L. monocytogenes, the first incorporating an IAC to be described for quantitative detection of a food-borne pathogen, is a simple and robust tool facilitating the identification of false negatives or underestimations of contamination loads due to PCR failure.
Many components of food products, culture media, and nucleic acid extraction reagents may inhibit PCR, leading to a dramatic decrease in sensitivity and even to false negative results (23, 26). In quantitative real-time (Q)-PCR, such inhibitors may cause underestimation of the contamination load in the sample, seriously compromising the applicability of this otherwise highly accurate technology (24). This is one of the major barriers to the systematic introduction of Q-PCR-based methods in routine food analysis. To tackle this problem, sample pretreatment procedures can be developed but, even if these are applied, it will always be necessary to assess PCR efficiency (or the performance of the sample pretreatment) in every reaction. The only way to achieve this is by the inclusion of an internal amplification control (IAC) (10, 20). A PCR IAC is a nontarget DNA fragment that is coamplified with the target sequence, ideally with the same primers used for the test reaction (6). In an IAC for Q-PCR, the forward and reverse target sequences are fused to both ends of a nontarget fragment, typically from an unrelated DNA, to which a second fluorescent probe (the IAC probe) hybridizes. The simultaneous use in a single reaction of two differently labeled fluorescent probes makes it possible to detect/quantify the target and to assess PCR efficiency at the same time. If negative results are obtained for the target PCR, the absence of a positive IAC signal indicates that amplification has failed (11).
A number of Q-PCR assays have been developed for the detection of food-borne pathogens, but few include an IAC. In particular, no IAC-containing assay has ever been developed for quantitative microbiological food analysis despite the generalized view that an IAC should be mandatory for PCR-based diagnostic tests (10). We report here the development and optimization of a novel Q-PCR assay for L. monocytogenes based on the simultaneous detection of hly gene target sequences, which we have shown to provide high specificity, sensitivity, and quantifiability (18), and an IAC sequence for the assessment of PCR inhibition. Using this assay, we show that some broth media widely used in the detection and enumeration of L. monocytogenes and certain food products commonly contaminated with these bacteria contain inhibitors that affect the analytical performance of the PCR.
IAC design and construction.
The IAC consisted of a 104-bp DNA fragment containing a portion of the acetyl-coenzyme A carboxylase gene from rapeseed (Brassica napus), BnACCg8 (GenBank accession no. X77576), flanked by the L. monocytogenes-specific hly gene sequences targeted by the previously described hlyQF and -R primers (18). This chimeric DNA fragment was generated by two rounds of PCR. The first used as template 100 ng of B. napus DNA and primers hlyAccF (5′-CATGGCACCACCAGCATCTGGTGAGCTGTATAATC) and hlyAccR (5′-ATCCGCGTGTTTCTTTTCGAGGCGCAGCATC), which contained the corresponding BnACCg8 target sequences plus a 5′ tail with the hlyQF/R primer sequences. The second PCR round used the purified first-round PCR product (diluted 1:1,000) as a template and the hlyQF/R primers. PCR conditions were as previously described (9). The IAC PCR product was purified, quantified using PicoGreen (Molecular Probes, Eugene, OR) in a luminescence spectrometer LS50B (Perkin-Elmer, Norwalk, CT), and diluted to the working concentration in double-distilled water containing 5 ng/μl tRNA as a blocking agent (to avoid binding of the negatively charged IAC DNA to the plastic microtubes).
With the exception of the BnACCg8 sequence (nucleotide positions 9651 to 9755), the IAC did not show significant similarity to any DNA sequence deposited in public DNA databases, as shown by BLAST-N searches (National Center for Biotechnology Information, Bethesda, MD; http://www.ncbi.nlm.nih.gov). The IAC and hly amplicons are specifically detected with previously described VIC- (8) and 6-carboxyfluorescein (FAM)-labeled (18) TaqMan probes, respectively. The IAC amplicon, 143 bp, is longer than the 64-bp hly-specific amplicon (18), facilitating distinction between these two PCR products by gel electrophoresis.
Optimization of hly-IAC Q-PCR assay.
The optimal IAC probe concentration (3, 21) was determined by performing Q-PCRs in the presence of 1,000 IAC molecules, no L. monocytogenes DNA, 100 nM FAM-labeled hly probe, and various amounts (from 25 to 250 nM) of the VIC-labeled IAC probe. The PCR conditions were those previously established for the hly-specific assay (18). The minimum probe concentration not resulting in an increase in cycle threshold (CT) was 100 nM. An excess of IAC may inhibit the target-specific reaction (5). To determine the optimal IAC concentration, we first performed Q-PCRs in the presence of various IAC amounts (1,000, 300, 100, 30, and 10 molecules per reaction) to determine the minimum required to give positive amplification. Ten IAC molecules were consistently detected, but the variation in VIC CT values was excessive (standard deviation [SD], >1.0). We then tested the three next lowest IAC amounts (30, 100, and 300 molecules) in the presence of L. monocytogenes CTC1010 (18) DNA corresponding to the quantification limit of the hly assay, previously determined to be 30 genome equivalents (GE) (note that the hly gene is in monocopy in the L. monocytogenes genome so that 1 GE corresponds to 1 bacterium or CFU in stationary phase) (16). The maximum IAC amount with no inhibitory effect on the hly-specific FAM signal was established at 100 copies.
Specificity and sensitivity of the hly-IAC Q-PCR assay.
We evaluated the specificity of the assay with 1 ng of genomic DNA (purified using the Wizard genomic DNA purification kit [Promega, Madison] and quantified with PicoGreen as above) from each of 49 L. monocytogenes strains, including representative strains of the different serovars of the species, and 96 nontarget bacteria, including 51 Listeria strains (17 L. innocua, 7 L. grayi, 10 L. seeligeri, 5 L. welshimeri, and 12 L. ivanovii) and 45 non-Listeria strains. The complete list of strains used can be found in Tables 1 and 2 of reference 18. The hly-IAC Q-PCR unequivocally distinguished L. monocytogenes isolates from nontarget bacteria. All reactions generated a positive IAC (VIC) signal, indicating that the lack of hly (FAM) signal that was obtained with non-L. monocytogenes isolates was not due to failure of the PCR.
TABLE 1.
Detection and quantification limits of the hly-IAC Q-PCR assaya
| Approx. no. of L. monocytogenes DNA molecules/reaction | Confidence interval limitb
|
hly system (FAM)
|
IAC system (VIC)
|
||||
|---|---|---|---|---|---|---|---|
| Lower | Upper | Signal ratioc | CTd | ΔRne | CTf | ΔRn | |
| 3 × 104 | 29,661 | 30,340 | 9 | 22.38 ± 0.11 | 0.98 ± 0.03 | 33.67 ± 0.79 | 0.10 ± 0.01 |
| 3 × 103 | 2,893 | 3,108 | 9 | 25.91 ± 0.10 | 0.92 ± 0.09 | 33.56 ± 0.75 | 0.12 ± 0.03 |
| 3 × 102 | 267 | 334 | 9 | 30.17 ± 0.15 | 0.91 ± 0.02 | 33.67 ± 0.57 | 0.26 ± 0.04 |
| 60 | 45 | 76 | 9 | 32.49 ± 0.19 | 0.81 ± 0.02 | 33.14 ± 0.47 | 0.43 ± 0.04 |
| 30 | 20 | 41 | 9 | 34.25 ± 0.16 | 0.78 ± 0.04 | 33.57 ± 0.66 | 0.53 ± 0.06 |
| 15 | 8 | 23 | 9 | 35.57 ± 0.57 | 0.75 ± 0.06 | 33.76 ± 0.74 | 0.65 ± 0.10 |
| 8 | 3 | 13 | 9 | 36.07 ± 0.59 | 0.73 ± 0.02 | 33.54 ± 0.47 | 0.70 ± 0.06 |
| 4 | 1 | 8 | 5 | 35.58 ± 0.76g | 0.79 ± 0.05 | 33.36 ± 0.31 | 0.73 ± 0.09 |
| 1 | 0 | 3 | 4 | 35.56 ± 1.03g | 0.85 ± 0.06 | 33.71 ± 0.75 | 0.70 ± 0.11 |
Note that both the hly and IAC templates are amplified by the same primers and that the number of copies of the hly target is variable, whereas that of the IAC template is constant (100 copies). This is reflected in the IAC data, in which the CT values remain constant, whereas the VIC fluorescence endpoints (ΔRn values) gradually decrease with increasing numbers of L. monocytogenes DNA molecules in the reaction. See Fig. 1 for representative amplification profiles for hly and IAC.
Calculated for the expected number of template molecules at each dilution with P as 0.05. The calculations were performed assuming a binomial distribution and confirmed by Monte Carlo simulations as previously described (18).
Signal ratio means positive reactions respective to nine reactions.
Cycle number at which fluorescence intensity equals a fixed threshold. FAM CT values were calculated with a prefixed threshold at 0.035 and a baseline from cycles 3 to 15.
ΔRn is the difference between R+n (reporter emission intensity/passive reference emission intensity) and R−n (background reporter emission intensity/passive reference emission intensity [calculated in no. template controls]) (3).
VIC CT values were calculated with a prefixed threshold at 0.035 and a baseline from cycles 3 to 23.
hly-negative amplifications were excluded from mean and SD calculations.
TABLE 2.
Performance of the hly-IAC Q-PCR with various media commonly used for Listeria
| Medium | Value obtained with:
|
||||
|---|---|---|---|---|---|
|
hly system (FAM)
|
IAC system (VIC)
|
||||
| CTa | ΔRnb | Relative accuracyc | CTd | ΔRnb | |
| Double-distilled water | 30.15 ± 0.15 | 0.95 ± 0.02 | 94.76 | 33.55 ± 0.61 | 0.55 ± 0.26 |
| BPW | 30.05 ± 0.16 | 0.98 ± 0.03 | 100.50 | 33.95 ± 0.52 | 0.60 ± 0.19 |
| BHI | 30.22 ± 0.12 | 0.96 ± 0.02 | 90.94 | 33.79 ± 0.49 | 0.65 ± 0.22 |
| Half-Fraser | 36.55 ± 1.05 | 0.70 ± 0.05 | 2.20 | 38.20 ± 1.20 | 0.13 ± 0.04 |
| Fraser | 37.25 ± 1.20 | 0.72 ± 0.05 | 1.45 | 38.52 ± 1.05 | 0.11 ± 0.04 |
Cycle number at which fluorescence intensity equals a fixed threshold. FAM CT values (mean plus or minus standard deviation) were calculated with a prefixed threshold at 0.035 and a baseline from cycles 3 to 15.
ΔRn is the difference between R+n (reporter emission intensity/passive reference emission intensity) and R−n (background reporter emission intensity/passive reference emission intensity [calculated in number of template controls]) (3).
Degree of correspondence between the response obtained by the reference method (2) and the response obtained by the alternative (Q-PCR) method.
VIC CT values were calculated with a prefixed threshold at 0.035 and a baseline from cycles 3 to 23.
To ensure maximum analytical sensitivity, the L. monocytogenes-specific signal should not be inhibited by the simultaneous coamplification of the IAC, particularly if small numbers of target molecules are expected. The detection limit of the hly-IAC assay was assessed by conducting Q-PCRs in the presence of 100 molecules of IAC and various amounts of genomic DNA from L. monocytogenes CTC1010 (equivalent to approximately 30, 15, 8, 4, and 1 GE per reaction). Table 1 shows FAM (hly) and VIC (IAC) CT and ΔRn values obtained in a total of nine replicates of three independent experiments. The Q-PCR assay detected as few as eight L. monocytogenes DNA molecules in 100% of the replicates and one to four target molecules in at least four out of the nine replicates. These results are similar to those previously reported for hly-specific uniplex assays (12, 16, 18). The IAC was coamplified in all reactions with overall CT values of 33.59 ± 0.68 and ΔRn values of 0.66 ± 0.11. Thus, the addition of 100 initial IAC molecules to the PCR mixture did not markedly decrease the sensitivity of the assay.
Quantifiability of the hly-IAC Q-PCR assay.
The capacity of the Q-PCR method to determine accurately the number of targets present in the sample depends upon the linearity and efficiency of the PCR. Linearity is the ability of the method to generate results proportional to the amount of analyte present in the sample and is represented by the regression coefficient. Efficiency is the capacity of the PCR to duplicate the amplicon molecules in each cycle and is calculated from the slope of the linear regression curve (s) from the equation E = 10−1/s−1 (14). These two parameters were assessed by carrying out PCRs with decreasing amounts of L. monocytogenes CTC1010 genomic DNA (equivalent to 3 × 104, 3 × 103, 3 × 102, 60, and 30 target DNA molecules per reaction). Figure 1 shows the typical amplification profiles obtained for each template. Table 1 shows FAM (hly) and VIC (IAC) CT and ΔRn values for nine replicates of three independent experiments.
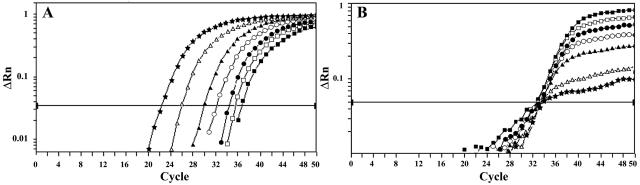
FIG. 1.
Representative amplification plots for hly (A) and IAC (B) templates obtained in the experiments shown in Table 1. Each reaction contained 100 IAC molecules and decreasing amounts of L. monocytogenes CTC1010 genomic DNA, equivalent to 3 × 104 (★), 3 × 103 (▵), 3 × 102 (▴), 60 (○), 30 (•), 15 (□), and 8 (▪) target molecules.
The relationship between the initial number of L. monocytogenes DNA molecules and FAM CT values was linear down to 30 target molecules, as indicated by the regression coefficient obtained (R2 = 0.997). At optimal efficiency (E = 1.00), the slope is −3.322 (15). The calculated slope for our hly-IAC PCR assays, −3.916, corresponds to an E value of 0.80, only slightly lower (12.6%) than that previously obtained for the uniplex hly assay (0.916) (18). These data, together with the small SD values for both replicates and independent experiments (Table 1), indicate that our hly-IAC Q-PCR assay accurately quantifies L. monocytogenes. The experimental quantification limit of the assay, 30 GE, coincided with the theoretical limit. The theoretical quantification limit was determined through the calculation of the expected number of template molecules at each dilution with the P value as 0.05 (the calculations were performed assuming a binomial distribution and confirmed by Monte Carlo simulations) and establishing as the theoretical quantification limit the lowest sample dilution in which the 95% confidence interval does not overlap with that of the next dilution (Table 1). This value is identical to that previously reported for the corresponding uniplex assay (18) and similar to that reported for other quantitative Q-PCR systems (4, 12, 13, 16, 21).
Performance of the hly-IAC assay.
The capacity of our assay to detect PCR inhibition was tested using four different broths typically employed for the culture, detection, or counting of L. monocytogenes: brain-heart infusion (BHI), buffered peptone water (BPW) (2), Fraser medium, and half-Fraser medium (7). The last two of these media are specified in ISO norms as enrichment media for the detection of L. monocytogenes in foodstuffs (1) and have been reported to inhibit PCR (23). We added 1 μl of broth medium or double-distilled water (control) to the standard hly-IAC Q-PCR mix containing 300 copies of genomic DNA from L. monocytogenes CTC1010.
The FAM (hly) and VIC (IAC) CT values obtained in the presence of BHI and BPW were similar to those for the control (P > 0.001) (Table 2). A mean of 287.16 ± 20.29 L. monocytogenes DNA molecules was detected on the basis of FAM CT values (95.72 ± 6.76%, quantification accuracy), with no inhibition of PCR, as shown by VIC CT values. In contrast, reactions containing Fraser or half-Fraser medium gave CT values that were significantly higher (P < 0.001) than those for the controls for both FAM and VIC signals, indicating that these media do indeed inhibit PCR. Significantly, although the hly target was amplified, the estimated number of copies, based on CT values, was below the quantification limit. Thus, in the absence of the corresponding IAC amplification profile, an underestimation by more than 2 orders of magnitude of the listerial contamination load would have passed unnoticed.
We also assessed the performance of the hly-IAC Q-PCR assay using foods in which L. monocytogenes is frequently found (25). Twenty-five-gram samples of raw pork meat, fermented pork sausage, cooked ham, frankfurter sausage, and raw or cold-smoked salmon were artificially contaminated with various amounts (approximately 3 × 107, 3 × 106, and 3 × 105 CFU/g) of L. monocytogenes CTC1010, as previously described (19, 22). These relatively high bacterial loads were used to enable accurate determination of the impact and scale of PCR inhibition on L. monocytogenes detection and quantification (something that would have been impossible with low bacterial numbers). The contaminated samples were immediately homogenized 1:10 (wt/vol) in BPW, and 1 μl of the homogenate was added to the standard hly-IAC Q-PCR mixture. In parallel, the number of L. monocytogenes CFU present in the samples was determined by standard plate counting (2). The results obtained are shown in Table 3.
TABLE 3.
Detection of PCR-inhibitory activity in different food matrices using the L. monocytogenes hly-IAC Q-PCR assaya
| L. monocytogenes contamination (CFU/g) | CFU/reaction | Results for:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fermented pork sausage
|
Frankfurter sausage
|
Cooked ham
|
Raw pork meat
|
Raw salmon
|
Smoked salmon
|
||||||||||||||
|
hlye
|
IAC
|
hlyf
|
IAC
|
hlyg
|
IAC
|
hlyh
|
IAC
|
hlyh
|
IAC
|
hlyh
|
IAC
|
||||||||
| CT valuesb | Relative accuracyc | CT valuesd | CT values | Relative accuracy | CT values | CT values | Relative accuracy | CT values | CT values | Relative accuracy | CT values | CT values | Relative accuracy | CT values | CT values | Relative accuracy | CT values | ||
| 3 × 107 | 3 × 103 | 26.05 ± 0.30 | 94.34 | 33.75 ± 0.71 | 25.94 ± 0.23 | 103.98 | 33.42 ± 0.35 | 26.05 ± 0.30 | 105.64 | 33.45 ± 0.64 | ND | NA | ND | ND | NA | ND | ND | NA | ND |
| 3 × 106 | 3 × 102 | 29.26 ± 0.31 | 113.15 | 33.52 ± 0.45 | 29.68 ± 0.37 | 93.06 | 33.69 ± 0.54 | 29.43 ± 0.17 | 90.19 | 33.62 ± 0.69 | ND | NA | ND | ND | NA | ND | ND | NA | ND |
| 3 × 105 | 3 × 101 | 33.01 ± 0.62 | 94.98 | 33.65 ± 0.54 | 29.43 ± 0.59 | 104.67 | 33.55 ± 0.75 | 32.49 ± 0.57 | 106.33 | 33.81 ± 0.45 | ND | NA | ND | ND | NA | ND | ND | NA | ND |
NA, not applicable; ND, amplification not detected.
FAM CT values (mean plus or minus standard deviation) were calculated with a prefixed threshold at 0.035, and a baseline from cycles 3 to 15.
Degree of correspondence between the response obtained by the reference method (2) and the response obtained by the alternative (Q-PCR) method. Note that in those samples where there was PCR inhibition as detected by the absence of IAC signal the relative accuracy values dropped dramatically.
VIC CT values (mean plus or minus standard deviation) were calculated with a prefixed threshold at 0.035, and a baseline from cycles 3 to 15.
Efficiency, 0.94; linearity, 0.9981.
Efficiency, 0.93; linearity, 0.9983.
Efficiency, 1.04; linearity, 0.9991.
Efficiency, not applicable; linearity, not applicable.
The FAM and VIC CT values obtained for fermented pork sausage, cooked ham, and frankfurter sausage samples were very similar (P > 0. 001) to those obtained with purified DNA (Tables 1 and 3), indicating that our hly-IAC Q-PCR system accurately detects and quantifies L. monocytogenes DNA in processed meat products. However, the L. monocytogenes-specific hly (FAM) signal was not detected in any of the raw pork meat and raw or cold-smoked salmon samples. This lack of FAM signal was accompanied by a lack of IAC (VIC) signal, indicating that the failure to detect L. monocytogenes DNA was a false negative result due to inhibition of the PCR.
Conclusions.
We have developed a Q-PCR assay with an IAC to facilitate monitoring of PCR inhibition and thus the identification of false negative results or target DNA underestimation due to PCR failure. This assay presents the same specificity, sensitivity, and quantification characteristics as the uniplex assay, demonstrating that the inclusion of an IAC does not compromise Q-PCR performance. The application of this assay to samples containing various broth media or food matrices relevant to Listeria demonstrated the presence of PCR inhibitors in some of these. Our data indicate that the hly-IAC Q-PCR assay here reported is a robust technique that can be routinely applied to the direct detection and quantification of L. monocytogenes DNA in food products.
Acknowledgments
We thank Marta Hugas and Nigel Cook for providing bacterial strains and DNA.
This work was supported by Spanish Ministerio de Ciencia y Tecnología grants AGL2002-03496 and BMC2000-0553. D.R.-L. was supported by fellowships from Universitat de Girona and the Leverhulme scheme of the Institute of Advanced Studies of the University of Bristol.
REFERENCES
- 1.Anonymous. 1996. Microbiology of food and animal feeding stuffs—Horizontal method for the detection and enumeration of Listeria monocytogenes—Part 1. Detection method (ISO 11290-1:1996). International Organization for Standardization, Geneva, Switzerland.
- 2.Anonymous. 1998. Microbiology of food and animal feeding stuffs—Horizontal method for the detection and enumeration of Listeria monocytogenes—Part 2. Enumeration method (ISO 11290-2:1998). International Organization for Standardization, Geneva, Switzerland.
- 3.Applied Biosystems. 1998. User bulletin #5, ABI Prism 7700 sequence detection system. Applied Biosystems, Foster City, Calif.
- 4.Bach, H. J., I. Jessen, M. Schloter, and J. C. Munch. 2003. A TaqMan-PCR protocol for quantification and differentiation of the phytopathogenic Clavibacter michiganensis subspecies. J. Microbiol. Methods 52:85-91. [DOI] [PubMed] [Google Scholar]
- 5.Ballagi-Pordány, A., and S. Belák. 1996. The use of mimics as internal standards to avoid false negatives in diagnostic PCR. Mol. Cell. Probes 10:159-164. [DOI] [PubMed] [Google Scholar]
- 6.Cone, R. W., A. C. Hobson, and M. L. Huang. 1992. Coamplified positive control detects inhibition of polymerase chain reactions. J. Clin. Microbiol. 30:3185-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser, J. A., and W. H. Sperber. 1988. Rapid detection of Listeria spp. in food and environmental samples by esculin hydrolysis. J. Food Prot. 51:762-765. [DOI] [PubMed] [Google Scholar]
- 8.Hernández, M., A. Rio, T. Esteve, S. Prat, and M. Pla. 2001. A rapeseed-specific gene, acetyl-CoA carboxylase, can be used as a reference for qualitative and real-time quantitative PCR detection of transgenes from mixed food samples. J. Agric. Food Chem. 49:3622-3627. [DOI] [PubMed] [Google Scholar]
- 9.Hoorfar, J., P. Ahrens, and P. Rådström. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoorfar, J., N. Cook, B. Malorny, P. Rådström, D. De Medici, A. Abdulmawjood, and P. Fach. 2003. Making internal amplification control mandatory for diagnostic PCR. J. Clin. Microbiol. 41:5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification control for diagnostic PCR assays. J. Clin. Microbiol. 42:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hough, A. J., S. A. Harbison, M. G. Savill, L. D. Melton, and G. Fletcher. 2002. Rapid enumeration of Listeria monocytogenes in artificially contaminated cabbage using real-time polymerase chain reaction. J. Food Prot. 65:1329-1332. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman, G. E., G. M. Blackstone, M. C. Vickery, A. K. Bej, J. Bowers, M. D. Bowen, R. F. Meyer, and A. DePaola. 2004. Real-time PCR quantification of Vibrio parahaemolyticus in oysters using an alternative matrix. J. Food Prot. 67:2424-2429. [DOI] [PubMed] [Google Scholar]
- 14.Klein, D., P. Janda, R. Steinborn, M. Muller, B. Salmons, and W. H. Gunzburg. 1999. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis 20:291-299. [DOI] [PubMed] [Google Scholar]
- 15.Knutsson, R., C. Löfström, H. Grage, J. Hoorfar, and P. Rådström. 2002. Modeling of 5′ nuclease real-time responses for optimization of a high-throughput enrichment PCR procedure for Salmonella enterica. J. Clin. Microbiol. 40:50-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nogva, H. K., K. Rudi, K. Naterstad, A. Holck, and D. Lillehaug. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl. Environ. Microbiol. 66:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Rodríguez-Lázaro, D., M. Hernández, M. Scortti, T. Esteve, J. A. Vázquez-Boland, and M. Pla. 2004. Quantitative detection of Listeria monocytogenes and Listeria innocua by real-time PCR: assessment of hly, iap, and lin02483 targets and AmpliFluor technology. Appl. Environ. Microbiol. 70:1366-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodríguez-Lázaro, D., A. Jofré, T. Aymerich, M. Hugas, and M. Pla. 2004. Rapid quantitative detection of Listeria monocytogenes in meat products by real-time PCR. Appl. Environ. Microbiol. 70:6299-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodríguez-Lázaro, D., M. D'Agostino, M. Pla, and N. Cook. 2004. A construction strategy for an internal amplification control (IAC) for molecular beacon-based real-time nucleic acid sequence-based amplification (NASBA). J. Clin. Microbiol. 42:5832-5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez-Lázaro, D., M. D'Agostino, A. Herrewegh, M. Pla, N. Cook, and J. Ikonomopoulos. 2005. Real-time PCR-based methods for quantitative detection of Mycobacterium avium subsp. paratuberculosis in water and milk. Int. J. Food Microbiol. 101:93-104. [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez-Lázaro, D., A. Jofré, T. Aymerich, M. Garriga, and M. Pla. Rapid quantitative detection of Listeria monocytogenes in salmon products: evaluation of pre-real-time PCR strategies. J. Food Prot. 68:1467-1471. [DOI] [PubMed]
- 23.Rossen, L., P. Nøskov, K. Holmstrøm, and O. F. Rasmussen. 1992. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA extraction solution. Int. J. Food Microbiol. 17:37-45. [DOI] [PubMed] [Google Scholar]
- 24.Scheu, P. M., K. Berghof, and U. Stahl. 1998. Detection of pathogenic and spoilage microorganisms in food with the polymerase chain reaction. Food Microbiol. 15:13-31. [Google Scholar]
- 25.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, W., B. González-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]