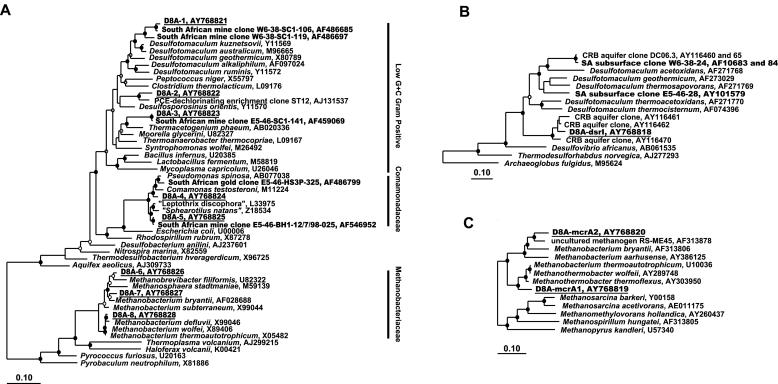
FIG. 3.
Phylogenetic analyses. Clones from Driefontein Mine boreholes are depicted with bold lettering (47). Those specific for this study are underlined. (A) Phylogenetic dendrogram based on a maximum likelihood analysis (Phylip FastDnaMl) of 862 unambiguously aligned positions of 16S rRNA gene sequences. (B) Maximum likelihood tree of dissimilatory sulfite reductase (dsrAB) genes. (C) Phylogenetic tree based on neighbor-joining analyses of methyl-coenzyme M (mcrA) reductase gene sequences. Maximum likelihood tree topologies were compared against trees generated by Bayesian inferences and parsimony. Closed circles indicate nodes conserved in all three treatments, with Bayesian probability and parsimony bootstrap values of >75%. Nodes conserved in the majority of tree topologies and with 50 to 74% probabilities and bootstraps are designated by open circles. Branch points supported by fewer than two trees or having <50% probability and bootstrap values have no designation. In the neighbor-joining tree, branch points supported by >79% bootstrap values are designated by solid circles, those supported by 60 to 79% bootstrap values are designated with open circles, and those having bootstrap values of <60% are unlabeled. Bars, changes per nucleotide.