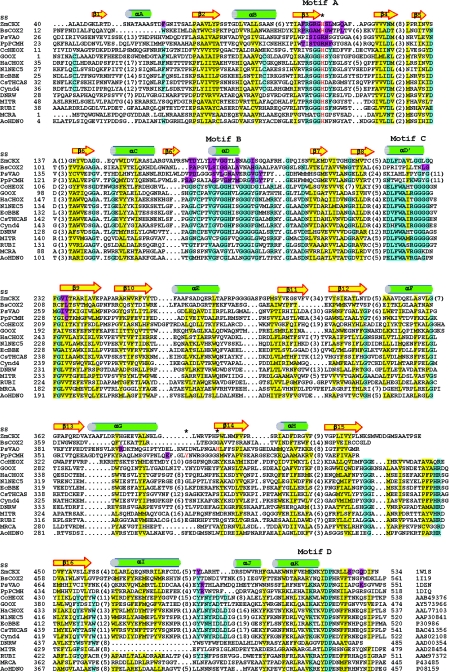
FIG.2.
Structure-based sequence alignment. The secondary structure elements for ZmCKX are labeled (SS). The protein accession numbers are indicated at the bottom on the right. The numbers of residues present in gaps are indicated in parentheses. The FAD-interacting residues in the known structures are indicted by magenta shading, while residues in the conservative hydrophobic core are indicated by yellow shading. There are four conserved segments in GOOX and related flavoproteins (motifs A to D), and the conserved residues are indicated by cyan shading. The flavinylation sites, Y384 in PpPCMH, H422 in PsVAO, and the consensus histidine in motif A, such as H105 in ZmCKX and H121 in BsCOX2, are indicated by red type and by asterisks. DNRW, MITR, RUBI, and MCRA are involved in the biosynthesis of antitumor antibiotics in Streptomyces; however, their substrates are still not known. Alignment was performed by manual editing based on the structural information and secondary structure prediction. This alignment suggests that the sugar oxidases, including GOOX, CcHEOX, NlNEC5, and HaCHOX, apparently evolved so that they have similar residues for FAD recognition but dissimilar residues for carbohydrate binding.