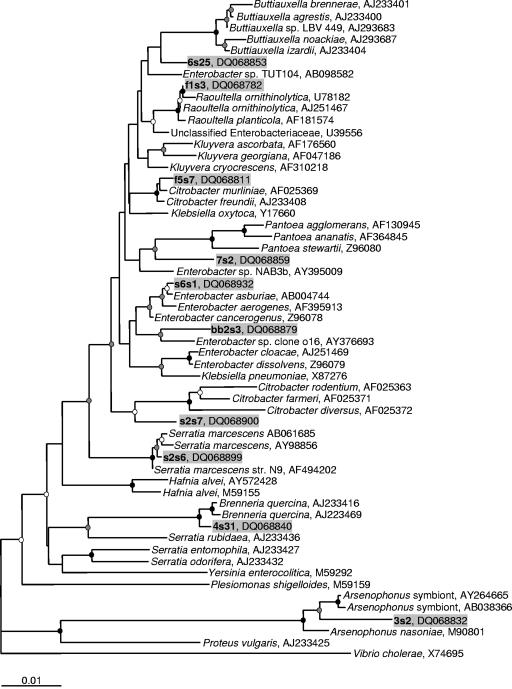
FIG. 2.
Inferred phylogenetic tree based on 16S rRNA gene sequences of representative clones belonging to the Enterobacteriaceae (OTUs 3 to 10, 12, and 13), including selected database sequences. The tree was generated using the neighbor-joining method with 2,000 bootstrap replicates using 1,416 nucleotides. Maximum-likelihood analysis resulted in similar tree topology (data not shown). Scientific names represent Bergey's classification from the Ribosomal Database Project-II website (15) and are followed by the GenBank accession number. Representative clones are listed in boldface type followed by GenBank accession numbers and include background shading. Branch points supported by bootstrap values of >90% are represented by black circles, >70% are represented by gray circles, and >50% are represented by white circles. Branches without circles are unresolved (bootstrap values of <50%). Database sequences included were chosen using the SeqMatch tool from the Ribosomal Database Project-II (15) with the addition of other sequences representing genera belonging to the Enterobacteriaceae. The tree was rooted with a 16S rRNA gene sequence from Vibrio cholerae, and the use of multiple different outgroup sequences from other Proteobacteria did not affect tree topology. The bar represents 0.01 substitutions per site.