Abstract
The luxS gene of Lactobacillus reuteri 100-23C was amplified by PCR, cloned, and then sequenced. To define a physiological and ecological role for the luxS gene in L. reuteri 100-23C, a luxS mutant was constructed by insertional mutagenesis. The luxS mutant did not produce autoinducers AI-2 or AI-3. Complementation of the luxS mutation by a plasmid construct containing luxS restored AI-2 and AI-3 synthesis. In vitro experiments revealed that neither the growth rate, nor the cell yield, nor cell survival in the stationary phase were compromised in the luxS mutant relative to the wild type and complemented mutant. The ATP content of exponentially growing cells of the luxS mutant was, however, 65% of that of wild-type cells. Biofilms formed by the luxS mutant on plastic surfaces in a bioreactor were thicker than those formed by the wild type. Biofilm thickness was not restored to wild-type values by the addition of purified AI-2 to the culture medium. In vivo experiments, conducted with ex-Lactobacillus-free mice, showed that biofilms formed by the mutant strain on the epithelial surface of the forestomach were approximately twice as thick as those formed by the wild type. The ecological performance of the luxS mutant, when in competition with L. reuteri strain 100-93 in the mouse cecum, was reduced compared to that of a xylA mutant of 100-23C. These results demonstrate that LuxS influences important ecological attributes of L. reuteri 100-23C, the consequences of which are niche specific.
The proximal region of the gastrointestinal tracts of rodents, pigs, chickens, and horses is inhabited by a Lactobacillus population. These bacteria adhere to, and proliferate on, the surface of nonsecretory epithelia present in the forestomachs, pars oesophagea, crops, and nonglandular stomachs of mice and rats, pigs, chickens, and horses, respectively (7, 32, 39). These epithelial associations can be termed biofilms because the bacteria appear to be encased in carbohydrate material and layers of bacterial cells several cells thick can be demonstrated in stained cryosections on the surface of proximal gut epithelia (7, 24). Shed from these biofilms, lactobacilli can be detected throughout the gut of the animal, including in the large bowel (23).
Lactobacillus reuteri strain 100-23 is an autochthonous inhabitant of the rodent gut. Recent molecular biological studies have begun to unravel the molecular mechanisms that enable this bacterial strain to reside in the guts of mice (34, 35). These studies have shown that L. reuteri 100-23 and the murine gut together provide an excellent paradigm to study the ecologically important characteristics, at the molecular level, of a gram-positive bacterial species that can form an epithelium-associated biofilm and of coping with ecological competition in the large bowel environment (34, 35).
Several examples of the ability of bacterial cells to alert their kindred to increasing cell density (quorum sensing) have been described (17). Typically, bacterial cells produce a small extracellular signal molecule, the autoinducer, and simultaneously sense it at the cell surface. If the concentration of autoinducer exceeds a certain threshold, the expression of specific genes is affected which, in turn, influences ecological behavior. Although oligopeptides seem to be the most likely signaling molecules in gram-positive bacteria (29), autoinducer 2 (AI-2), associated with the expression of the luxS gene, may also be of interest. A wide range of gram-positive and gram-negative bacterial species produce AI-2 by a common biosynthetic pathway, and it has been proposed that AI-2 is a universal signaling molecule that functions in both intra- and interspecies cell-to-cell communication (26, 38). Recently, production of a further autoinducer (AI-3) has been associated with the luxS gene (9).
Influences of AI-2 on the formation of biofilms by gram-positive bacteria indigenous to the oral cavity have been reported (2, 15, 16). Although effects on ecological behavior of gut commensals do not appear to have yet been reported for AI-2, it might not be surprising if the members of gut communities were to somehow communicate among themselves and that AI-2 was a regulatory factor in determining community structure and function. Therefore, we have studied the influence of mutation of the luxS gene on the ability of L. reuteri 100-23 to form biofilms in vitro and in vivo, and to compete with other lactobacilli in the intestinal tracts of mice. We accomplished this work by comparing the characteristics and behavior of the wild-type strain 100-23C (a plasmid-free derivative of strain 100-23) with that of a derivative mutated in the luxS gene and thus unable to synthesize AI-2 or AI-3.
MATERIALS AND METHODS
Lactobacillus strains.
The Lactobacillus strains used in the present study are listed in Table 1.
TABLE 1.
Lactobacillus strains used in the study
| Lactobacillus strain | Species | Relevant characteristics |
|---|---|---|
| 100-23 | L. reuteri | Rodent gut isolate |
| 100-23C | L. reuteri | Plasmid-free derivative of 100-23 |
| 100-23C xylA | L. reuteri | 100-23C mutated in the xylose isomerase gene, xylA |
| 100-23C luxS | L. reuteri | 100-23C mutated in the luxS gene |
| 100-23 luxS (pGS1) | L. reuteri | 100-23C luxS complemented with plasmid pGS1 containing a copy of the luxS gene |
| 100-93 | L. reuteri | Rodent gut isolate |
| 100-5 | L. johnsonii | Rodent gut isolate |
Detection and characterization of the luxS gene of L. reuteri 100-23C.
An internal region (378 bp) of the L. reuteri 100-23C luxS gene was amplified by using the degenerate primers LuxSlacto(F) and LuxSLacto(R) (Table 2). These primers were constructed to target two conserved regions of the luxS genes of Lactobacillus gasseri ATCC 33323 (NCBI accession no. NZ_AAA002000003), Lactobacillus plantarum WCFS1 (NCBI accession AL935254), Oenococcus oeni (NCBI accession X82326), and Streptococcus pyogenes (NCBI accession no. AE014163). The PCR product was purified and cloned in Escherichia coli DH5α by using the pGEM-T Easy system (Promega, WI) and sequenced with primers T7 and SP6. Regions adjacent to this sequence were amplified by inverse PCR with the primer combinations Inverse2(F)-Int.luxS(R) and Inverse4(F)-Inverse1(R) and using TaqI digested and self-ligated chromosomal DNA of L. reuteri 100-23C. PCR products were purified and sequenced directly by using primers Int.luxS(R) and Inverse4(F), resulting in a TaqI fragment sequence of 1,051 bp containing the complete luxS gene and its putative promoter sequence. This fragment was amplified with primers LR+P(F) and LRluxS(R), cloned in pGEM-T/E. coli DH5α, and sequenced in order to confirm the sequence revealed by inverse PCR. The luxS gene sequence of strain 100-23C has been deposited as accession AY485153 in the NCBI databank.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′)a | Application |
|---|---|---|
| LuxSlacto(F) | AAAGTTTTRMATTAGATCATAC | Amplification of internal region of the luxS gene |
| LuxSlacto(R) | ATGGTCTYKGTAGTTMCCACA | Amplification of internal region of the luxS gene |
| Inverse2(F) | GGAAGATGTTCAGGGAACAAC | Inverse PCR |
| Inverse4(F) | TGCATACGATTGAACACTTGC | Inverse PCR |
| Inverse1(R) | GAACGTAAGGTGCCTTAACC | Inverse PCR |
| LR+P(F) | GAGCATAGTGTGATGATCC | Sequence confirmation |
| LRluxS(R) | CGAGAAATCCTTAGTCAGC | Sequence confirmation and probe |
| LRluxS(F) | TGGTTGGATTGCCGTTCC | Probe |
| Int.luxS(F) | GACTGGATCCTAAGTTAAGGCACCTACGTTC | Mutation |
| Int.luxS(R) | GACTGAATTCAGCCAAATGGAGAGCAATC | Mutation and inverse PCR |
| LR+Pcomp(F) | GACTGTCGACGAGCATAGTGTGATGATCC | Complementation |
| LR+Pcomp(R) | GACTGTCGACCGAGAAATCCTTAGTCAGC | Complementation |
Restriction sites are italicized; translation stop codons are in boldface.
Derivation of mutant and complemented strains.
A L. reuteri 100-23C strain in which the xylA gene was mutated by insertional mutagenesis was derived previously (35). Insertional mutagenesis of the luxS gene was achieved by site-specific integration of plasmid pORI28 into the L. reuteri 100-23C chromosome as described by Walter et al. (35) using the temperature sensitive plasmid pVE6007 (12) as the helper plasmid. An internal region of the luxS gene was amplified by PCR with the primers Int.luxS(F) and Int.luxS(R) and cloned into pORI28 by directional cloning as described by Russell and Klaenhammer (21), resulting in plasmid pILX104. To derive a probe, primers LRluxS(F) and LRluxS(R) were used to amplify a PCR product containing the complete luxS gene of L. reuteri 100-23C. The integration of pILX104 into the luxS gene was confirmed by PCR with primers flanking the target region and by Southern blotting using the LRluxS probe and standard methods (22). The in vitro stability of the integration was determined by serial subculture in Lactobacilli MRS medium in the absence of erythromycin as described previously (35).
The L. reuteri-E. coli shuttle vector p29cat232small (34) was used to construct a plasmid for complementation of the luxS mutant strain. The entire luxS gene and the putative promoter were amplified from L. reuteri 100-23C genomic DNA by using primers LR+Pcomp(F) and LR+Pcomp(R) (these primers contain SalI restriction sites) (Table 2). The PCR product was purified, digested with SalI, and cloned in SalI-digested, alkaline phosphatase-treated p29cat232small. The ligation mixture was used to electrotransform L. reuteri DSM 20016T (14). The resulting complementation plasmid, pGS1, was purified and used to electrotransform the L. reuteri 100-23C luxS mutant strain.
Detection of in vitro and in vivo AI-2 production by lactobacilli.
Aliquots of Lactobacillus cultures in Lactobacilli MRS medium (Difco; anaerobic, 37°C) were centrifuged (7,740 × g, 5°C, 5 min) and the supernatants were retained and filtered (0.22-μm pore size). Then, 1-ml volumes were freeze-dried for storage. Stomach contents collected from 6-week-old mice were pooled (five mice per pool), suspended in sterile water (10% [wt/vol]), and homogenized by using glass beads and vortex mixer. After centrifugation (as described above), the supernatants were filtered (0.22-μm pore size), and 1-ml volumes were freeze-dried for storage. For AI-2 assays, the freeze-dried supernatants were dissolved in one ml of methanol and mixed vigorously. Insoluble material was removed by centrifugation at 10,000 × g for 10 min. The supernatants were recovered, dried under vacuum, dissolved in 1 ml of distilled water, and then stored at −80°C.
AI-2 assays with the biosensor Vibrio harveyi MM32.
AI-2 activity was measured as described previously (1) but using the V. harveyi MM32 (ΔluxN ΔluxS) AI-2 biosensor strain kindly provided by Bonnie Bassler (Princeton University, New Jersey). Strain MM32 does not produce endogenous AI-2 but emits light proportionally to exogenous levels of AI-2 (5). Strain MM32 was grown overnight at 30°C in AB medium (0.3 M sodium chloride, 0.1 M magnesium sulfate, 2% [wt/vol] Casamino Acids [pH 7.5; potassium hydroxide], 0.01 M dipotassium phosphate, 1% [vol/vol] glycerol, 1 mM arginine) and diluted 1/5,000 in fresh AB medium. Luminescence was measured at 30°C in 96-well black plates with a luminometer (Victor2; Wallac) at 30-min intervals and recorded as counts per second. Each well contained a 100-μl volume of diluted MM32 culture supplemented with 2, 5, or 10% (vol/vol) of the test sample. Synthetic AI-2 prepared as described previously (25) was used as a positive control, and diluted MM32 culture provided a negative comparison. This assay does not detect AI-3 (28).
Preparation of AI-2 cell-free culture supernatant.
A concentrated AI-2 preparation for use in bioreactor experiments was prepared as osmotic shock fluid (31). One liter of Luria broth containing 0.5% glucose was inoculated with 10 ml of an overnight culture of Salmonella enterica serovar Typhimurium 14028. The culture was grown with shaking at 37°C for 4 h. The cells were harvested by centrifugation at 7,000 × g for 10 min at 20°C. The cell pellets were suspended in 400 ml of 0.4 M sodium chloride solution. The suspended cells were shaken at 37°C for 2 h and harvested by centrifugation at 7,000 × g for 10 min at 20°C, and the supernatant was filtered (0.22 μm). The cell-free supernatant was then lyophilized for storage. Activity was measured using the V. harveyi biosensor strain MM32: AI-2 activity was the same before and after lyophilization.
Detection of in vitro AI-3 production by isogenic strains of 100-23C.
AI-3 production was kindly determined by Vanessa Sperandio (University of Texas Southwestern Medical Center). The assay utilized a transcriptional fusion of the locus of enterocyte effacement (LEE) from E. coli O157:H7 with lacZ. AI-3 activates transcription of the LEE genes and hence LacZ, which can be measured in terms of β-galactosidase activity. E. coli TEVS232 (LEE1::lacZ) was grown in Dulbecco modified Eagle medium to an A600 of >2.0 at 37°C with rotation at 250 rpm, as described previously (27). Filtered (0.2-μm pore size) supernatant from 4- or 8-h cultures was added to the test cultures to give a final concentration of 10% (vol/vol). Each strain was tested in triplicate assays. This assay does not detect AI-2 (28).
Determination of bioenergetic parameters of isogenic strains of 100-23C.
The electrical potential across the cell membrane (Δψ) of logarithmic-growth-phase cells that were harvested by centrifugation (8,000 × g, 20 min, 4°C) and washed twice in 50 mM potassium phosphate buffer containing 5 mM MgCl2 (pH 7.5) was measured. The cells were energized by using 20 mM glucose for 15 min, followed by the addition of [3H]methyltriphenylphosphonium iodide (3HTPP+; 30 to 60 Ci mmol−1; 1 μM final concentration). [1,2-3H]taurine (50 μM; 5 to 30 Ci mmol−1) and 3water (25 mM; 25 mCi g−1) were used to determine the intracellular volume. Taurine was not metabolized by L. reuteri 100-23C. The radiochemicals were obtained from NEN (Life Science Products, Inc.). After incubation for 10 min at 37°C, the cultures were centrifuged through 350 μl of silicon oil (BDH Laboratory Supplies, Poole, England; 40% mixture of phthalic acid bis[2-ethyl-hexyl ester] and 60% silicone oil [mixture of 40% DC200/200 silicone oil and 60% DC 550] which had been left overnight in an anaerobic chamber at room temperature to equilibrate) in 1.5-ml microcentrifuge tubes (13,000 × g, 5 min, 22°C), and 20-μl samples of supernatant were removed. The tubes and contents were frozen (−20°C), and cell pellets were removed with dog nail clippers. Supernatant and cell pellets were added to scintillation fluid, and the radioactivity was measured by using a scintillation counter (LKB Wallac 1214 Rackbeta). The intracellular volume (3.0 ± 0.7 μl mg of protein−1) was estimated from the difference between the partitioning of 3water and [1,2-3H]taurine. Δψ was calculated from the uptake of [3H]TPP+ according to the Nernst relationship. Nonspecific TPP+ binding was estimated from cells that had been treated with valinomycin and nigericin (15 μM each) for 25 min. The ionophore nigericin mediates an electroneutral exchange between potassium and protons and, in combination with valinomycin, causes a complete dissipation of K+ andH+ gradients across the cell membrane. Protein from sodium hydroxide-hydrolyzed cells (0.2 M NaOH, 100°C, 20 min) was assayed by the method of Markwell et al. (13).
ATP was extracted from logarithmic-growth-phase cells by perchloric acid and sodium chloride-sodium bicarbonate treatment after separation of the cells from the growth medium, as previously described (3), and stored at −70°C. Prior to analysis, samples were thawed and the potassium perchlorate was removed by centrifugation (13,000 × g, 5 min, 22°C). The samples (50 μl) were then diluted in 400 μl of 40 mM Tris-acetate buffer (pH 7.75) containing 2 mM EDTA and 50 mM MgCl2. The luciferase reaction (10) was initiated by adding 50 μl of a purified luciferin-luciferase mix to 450 μl of the diluted extract according to the supplier's recommendations (Sigma). Light output was immediately measured with a luminometer (model LB935; Berthold) using an ATP standard. ATP (sodium salt), luciferin, d-luciferase (EC 1.13.12.7), valinomycin, and nigericin were obtained from Sigma.
Bacterial growth rates in vitro.
Volumes (200 ml) of Lactobacilli MRS medium were prereduced and prewarmed (37°C) in an anaerobic glovebox 24 h prior to use. Each volume was inoculated with 2 ml of overnight Lactobacillus culture, and 1-ml aliquots were removed at hourly intervals over 8 h to determine the A600 by using a spectrophotometer (Novospec II; Pharmacia Biotech). The growth rate constant (k) for the logarithmic phase of growth was determined by plotting the log10 of the optical density over time (19).
In vitro cell yield.
Ten-milliliter volumes of Lactobacilli MRS medium were inoculated with 10 ml of overnight culture of lactobacilli and incubated anaerobically at 37°C for 24 h (stationary-phase culture). Five replicate cultures were prepared per bacterial strain. The bacterial cells were harvested by centrifugation (7740 × g, 5°C, 5 min), the supernatant was carefully removed without disturbing the pelleted cells, and the cells were left to dry at 37°C until a constant weight was achieved.
Bacterial survival in vitro.
Ten-milliliter volumes of Lactobacilli MRS medium were inoculated with 10 μl of an overnight culture of lactobacilli and incubated anaerobically at 37°C for 72 h. Then, 1-ml aliquots were removed after 8 h of incubation, and thereafter each 24 h, serially diluted to 10−6 in phosphate-buffered saline (PBS), and colonies were counted on Lactobacilli MRS agar plates that had been spread plated with 100-μl volumes of each dilution. Percent survival relative to the CFU per ml obtained after 8 h of incubation was calculated from 24-, 48-, and 72-h CFU values. The minimum detection limit was 100 CFU/ml, and assays were conducted in triplicate.
In vitro biofilm formation by wild-type and luxS mutant strains.
A bioreactor (flow cell) containing 10 plastic (Thermanox; Nalge Nunc International) coupons (10.5 by 22 mm) suspended on wires at 10-mm intervals was assembled and sterilized by autoclaving. The reactor was filled with 100 ml of half-strength Lactobacilli MRS medium, sufficient to cover the coupons. One milliliter of overnight bacterial culture was added. The apparatus was maintained under a constant flow of nitrogen throughout the experiment. After an initial static period of 17 h, fresh medium was continuously added at a dilution rate of D = 1 h−1 for up to 72 h. The medium in the reactor was stirred during the experiment. The flow condition used was faster than the doubling rate of the lactobacilli under these conditions (t = 1.03 h−1), effectively selecting for cells in the bioreactor that could attach to and multiply on the coupon surface. Lactobacillus colonies associated with biofilms were counted on Lactobacilli MRS medium with or without the addition of 5 μg of erythromycin per ml of medium after removal from coupon surfaces using a sterile cotton swab. Each strain was tested on two separate occasions.
Biofilm formation by the luxS mutant strain in the presence of AI-2 was tested by adding osmotic shock fluid from serovar Typhimurium 14028 (prepared as described above) to the culture medium at a final concentration of 0.1%.
Coupons removed from the reactor were dipped in 0.1% (wt/vol) peptone solution to remove nonadherent cells and then placed in 10 ml of phosphate wash solution (PWS; 10 mM potassium dihydrogen phosphate, 0.85% sodium chloride, 5 mM magnesium chloride [pH 7.1]) for 2 min. The biofilm was fluorescently stained by placing the coupon in 10 ml of PWS containing acridine orange (1.25 mg ml−1) for 2 min. After a further 2 min in PWS, the biofilm was observed using a single-channel pass on a confocal laser scanning electron microscope (Zeiss 510 LSM). The biofilm was visualized by using a 488-nm excitation wavelength with emissions collected at a long-pass of 505 nm onwards. Fifteen measurements of biofilm depth (z-sections) were collected in a grid pattern across the coupon.
Experiments with mice.
Lactobacillus-free mice were inoculated by intragastric gavage (dose of 5 × 107 lactobacilli), as in our previous experiments to gauge the ecological performance of Lactobacillus strains (35). The mice were 6 weeks of age (males and females) at the time of inoculation, were maintained in isolators by gnotobiotic technology, and had originated from our colony of animals that has been described previously (33). The bacterial cells had been washed and suspended in PBS (pH 7.4) prior to use as an inoculum. The animals were killed, and samples were collected for bacteriological culture or other analysis 2 weeks after inoculation with lactobacilli. These experiments were conducted with approval from the University of Otago Animal Ethics Committee (approval number 104/02).
In vivo biofilm formation.
Measurements of biofilms formed on the epithelial surface of the murine forestomach were made 2 weeks after inoculation of Lactobacillus-free animals with pure cultures of the wild type or the mutant strain. The stomachs were carefully excised from each animal, slit lengthwise and the contents completely removed by gentle washing with PBS (pH 7.4). Unfixed stomachs were stained with 1 mM BacLight Green (Molecular Probes, Oregon), a bacterium-specific stain, according to the manufacturer's instructions, and examined with a laser scanning confocal microscope (Zeiss 510 LSM). Z sections (xz) of the bacterial layer were visualized using an argon laser's excitation at 488 nm and a 505-nm long-pass emission filter. Fifteen measurements of depth were obtained at approximately equidistant intervals along each section. The forestomach of a noncolonized lactobacillus-free mouse and a pure culture of wild-type L. reuteri 100-23C served as negative and positive controls, respectively.
Ecological competitiveness.
Measurements of ecological competitiveness of strains were made by inoculating Lactobacillus-free mice with 1:1 mixtures of mutant and wild-type strains (total dose of 5 × 107 lactobacilli). Quantification of total Lactobacillus populations in forestomach, jejunal, and cecal samples was achieved by using Rogosa SL agar (Difco) as described previously (35). Mutant colonies were counted on Rogosa SL agar plates containing 5 μg of erythromycin per ml of medium (35).
Statistical comparisons.
The Mann-Whitney nonparametric test was used to compare experimental results.
RESULTS
luxS gene of L. reuteri 100-23C.
Sequence analysis of the L. reuteri 100-23C luxS gene revealed a complete open reading frame (477 bp), a putative ribosomal binding site (AGGAGA-8 nucleotides-ATG), and a putative promoter (TTGACA-17 nucleotides-TATTATAAT). BlastP search revealed highest homologies to LuxS of Lactobacillus acidophilus, L. johnsonii, and Pediococcus pentosaceus (85, 85, and 82% identity, respectively). Southern analysis revealed that luxS was present as one gene copy in the genome of 100-23C. Growth of the luxS mutant strain in the absence of antibiotic showed that 96% of the population retained erythromycin resistance over 100 generations. This was similar to the in vitro stability of insertions described for insertional mutants of other lactobacilli, including other mutants of L. reuteri 100-23C (21, 35).
In vitro and in vivo production of AI-2 by lactobacilli.
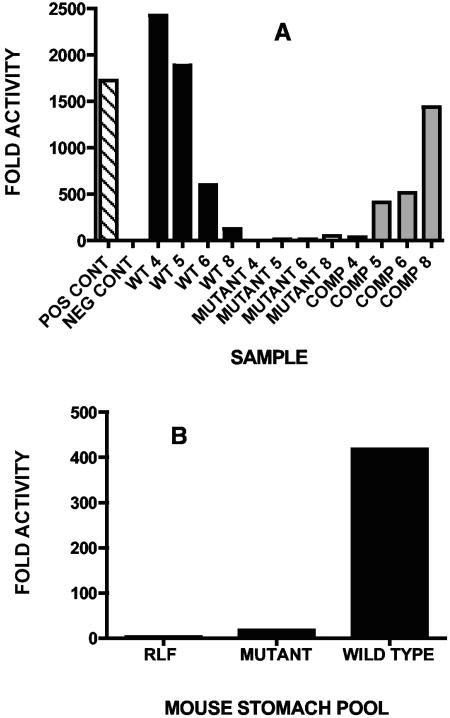
AI-2 activity was detected in supernatants of cultures obtained from the wild type and complemented mutant strains. Highest activity was detected in supernatant from 4-h (logarithmic growth) wild-type culture (Fig. 1A). In contrast, highest activity was detected in stationary-phase culture of the complemented mutant strain. This growth-phase-dependent accumulation of AI-2 activity has been reported for other bacteria (6, 11, 18, 30, 37). Four-hour cultures of the two Lactobacillus strains used as competitors in ecological performance studies contained detectable AI-2: L. reuteri 100-93, 4,715-fold activity relative to the negative control; L. johnsonii 100-5, 21-fold activity relative to the negative control.
FIG. 1.
(A) AI-2 activity (fold values relative to negative control) in Lactobacillus culture supernatants. Single assay per supernatant. Pos Cont, positive control, addition of synthetic AI-2; Neg Cont, negative control, biosensor only; WT, L. reuteri 100-23C; Mutant, L. reuteri 100-23C luxS mutant; Comp, L. reuteri 100-23C luxS mutant complemented by pGS1; 4, 5, 6, and 8, hours of culture incubation. (B) AI-2 activity (fold values relative to negative control) in mouse stomach preparations. Five stomach contents pooled per group. Single assay per mouse group. RLF, Lactobacillus-free mice; Mutant, mice colonized by L. reuteri 100-23C luxS mutant; Wild type, mice colonized by L. reuteri 100-23C.
The stomach contents of Lactobacillus-free mice did not contain detectable AI-2 activity, whereas samples from mice colonized by the wild-type strain contained readily detectable levels of AI-2 (Fig. 1B). Stomach contents from mice colonized by the luxS mutant strain had activity barely above background level. This activity could have resulted from fecal bacteria present in the stomach due to coprophagy.
In vitro AI-3 production by lactobacilli.
The luxS mutant did not produce detectable amounts of AI-3 activity, whereas the wild-type and complemented strains produced the autoinducer in stationary phase (mean 300.0 [standard error of mean 10.58]; 328.3 [56.1] Miller units, respectively, compared to the negative control 174.7 [33.4]) but not in logarithmic growth phase.
Physiological comparisons of the luxS mutant, wild-type, and complemented mutant strains.
The wild type, luxS mutant, and complemented strains did not differ in either their maximum specific growth rate (0.43 h−1) or cell yield (wild-type mean, 0.025 g [dry weight] of cells [standard error of mean = 0.001]; luxS mutant, 0.023 g [0.001]; complemented strain, 0.025 g [0.001]) in Lactobacilli MRS medium. The cell viability of stationary-phase cultures was similar for the mutant, wild-type, and complemented strains over a 72-h period by which time ca. 1.5% of the cells enumerated after 8 h of incubation could be recovered by culture.
The energy status of logarithmic-phase cells was assessed by measuring intracellular ATP content and the electrical potential (Δψ) across the cell membrane. The wild-type and luxS complemented strains contained more ATP per mg of protein during exponential growth (at identical optical densities) than those of the luxS mutant strain (mean and standard error of the mean of two assays = 1,338 ± 110 nmol, 1,326 ± 146 nmol, and 867 ± 78 nmol, respectively). The Δψ was in the range −100 to 115 mV for all three strains.
In vitro biofilm formation.
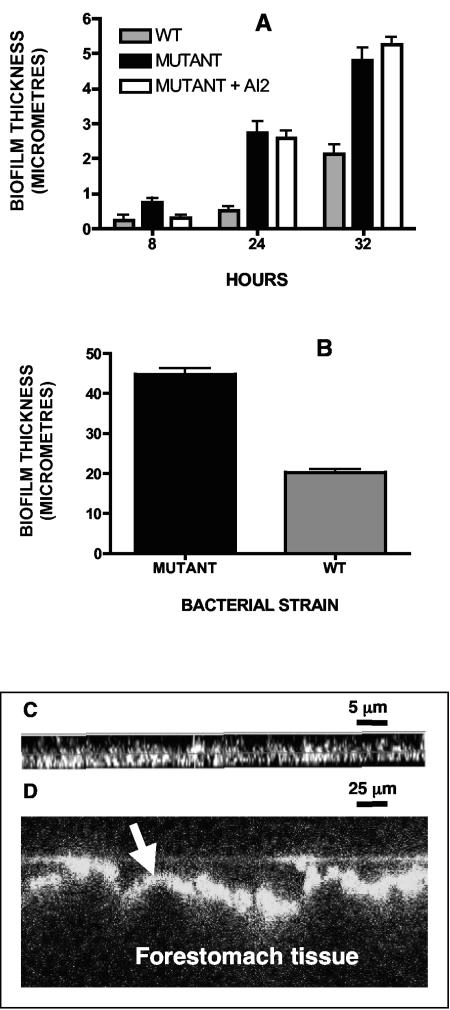
Measurement of biofilm thickness on plastic coupons that had been suspended in flow cells inoculated with lactobacilli was achieved by confocal laser microscopy. Maximum biofilm thickness was observed from 32 h onward after inoculation of the flow cell. The luxS mutant strain formed thicker biofilms than the wild-type strain (Mann-Whitney test, P < 0.0001) at all times at which measurements were made (Fig. 2A). Quantitative bacteriological culture showed that the number of Lactobacillus cells adhering to plastic coupons was the same for mutant and wild-type strains (ca. 4 × 108 per coupon). Addition of an AI-2 preparation to the bioreactor culture did not influence the thickness of the biofilm produced by the mutant (Mann-Whitney test, P > 0.05) except at 8 h (P = 0.0142). The 8-h result might have been due to the difficulty of measuring the very thin biofilms present at this stage of biofilm formation rather than to an effect of exogenous AI-2.
FIG. 2.
(A) Mean biofilm thickness (bars indicate the standard error of the mean) formed on plastic coupons in bioreactor by L. reuteri 100-23C strains. Thirty measurements collected during duplicate experiments for each strain per time interval. WT, wild-type strain; Mutant, luxS mutant strain; Mutant + AI2, luxS mutant strain with addition of AI-2 extracted from serovar Typhimurium 14028. (B) Mean biofilm thickness (bars are standard error of the mean) on forestomach epithelium of mice colonized by L. reuteri 100-23C strains; Mutant, mice colonized by luxS mutant strain; WT, mice colonized by wild-type strain. Seven mice per group, 105 measurements per mouse group. Mann-Whitney test P < 0.0001. (C and D) Examples of biofilm images obtained by confocal laser microscopy: in vitro biofilm on plastic surface (C) and in vivo biofilm on epithelial surface (D) (the arrow indicates fluorescent biofilm).
In vivo biofilm formation.
Measurement of Lactobacillus biofilms on the forestomach epithelium of ex-Lactobacillus-free mice by using confocal laser microscopy showed that the luxS mutant formed thicker biofilms than did the wild-type strain (Fig. 2B and C; Mann-Whitney test, P < 0.0001).
Ecological competitiveness of the luxS mutant in the gastrointestinal tracts of mice.
Total Lactobacillus populations in gut samples did not differ between groups inoculated with pure cultures of mutant or wild-type strains or with mixtures of lactobacilli (stomach, ∼108 per g; jejunum, ∼107 per g; cecum, ∼108 per g). The proportion of the total Lactobacillus population formed by the luxS mutant in the stomach and jejunum did not differ from that of the xylA mutant regardless of competitor strain (L. johnsonii 100-5 or L. reuteri 100-93; Table 3; Mann-Whitney test, P > 0.05). The proportion of the total population formed by the luxS mutant in the cecum differed to that of the xylA mutant when L. reuteri 100-93 was the competitor (Table 3; Mann-Whitney test, P = 0.0427). The luxS and xylA mutants were equally competitive in the cecum with regard to L. johnsonii 100-5 (Table 3; Mann-Whitney test, P > 0.05).
TABLE 3.
Mutant populations in ceca of mice
| Mouse inoculum | Organ | No. of mice examined | Mean % of total Lactobacillus population (SEM) |
|---|---|---|---|
| luxS mutant and L. johnsonii 100-5 | Stomach | 12 | 57.4 (9.9) |
| xylA mutant and L. johnsonii 100-5 | Stomach | 11 | 68.7 (6.5) |
| luxS mutant and L. johnsonii 100-5 | Jejunum | 12 | 67.8 (9.8) |
| xylA mutant and L. johnsonii 100-5 | Jejunum | 11 | 68.4 (8.4) |
| luxS mutant and L. johnsonii 100-5 | Cecum | 12 | 56.4 (11.1) |
| xylA mutant and L. johnsonii 100-5 | Cecum | 11 | 60.1 (8.7) |
| luxS mutant and L. reuteri 100-93 | Stomach | 15 | 32.4 (7.8) |
| xylA mutant and L. reuteri 100-93 | Stomach | 13 | 46.4 (8.6) |
| luxS mutant and L. reuteri 100-93 | Jejunum | 15 | 22.5 (3.9) |
| xylA mutant and L. reuteri 100-93 | Jejunum | 13 | 43.5 (10.3) |
| luxS mutant and L. reuteri 100-93 | Cecum | 15 | 15.0 (2.7) |
| xylA mutant and L. reuteri 100-93 | Cecum | 13 | 43.1 (8.7) |
DISCUSSION
Orthologs of the luxS gene have been detected in a wide range of gram-positive and gram-negative bacterial species, and it has been proposed that AI-2 is a universal signaling molecule that functions in interspecies cell-to-cell communication (25). AI-2 is produced from S-adenosylmethionine (SAM) in three enzymatic steps (26). The use of SAM as a methyl donor in metabolic processes such as DNA synthesis produces a toxic intermediate S-adenosylhomocysteine (an inhibitor of the SAM-dependent methyltransferases) that is hydrolyzed to S-riboadenosylhomocysteine (SRH) by a nucleosidase. The protein LuxS, encoded by the luxS gene, catalyzes the cleavage of SRH to form homocysteine and 4,5-dihydroxy 2,3-pentanedione. The latter molecule cyclizes to form pro-AI-2 that is then transformed, at least in the case of V. harveyi, to AI-2 by the addition of boron (25).
Our investigation of a strain of L. reuteri, autochthonous to the gastrointestinal tract of mice, has shown that LuxS is essential for the synthesis of both AI-2 and AI-3 by these bacteria. Complementation of the luxS mutation using a plasmid containing a wild-type copy of luxS restored production of AI-2 and AI-3, demonstrating that the loss of AI-2 and/or AI-3 production was due to inactivation of the luxS gene and not to a polar effect. Much less is known about AI-3 compared to AI-2; therefore, we have concentrated on the latter substance to provide evidence of LuxS function under both in vitro and in vivo conditions. Minimal AI-2 activity was detected in the stomachs of Lactobacillus-free mice, whereas colonization of this site by wild-type 100-23C resulted in detection of the autoinducer at 400-fold-higher levels.
Biofilm thickness, both in vitro and in vivo, was increased in the case of the luxS mutant strain. This could equally be due to the loss of quorum-sensing function and therefore loss of regulatory constraint on Lactobacillus proliferation (38) or the loss of a means of recognizing the boundaries of the habitat (20). It is more difficult to envisage the increased biofilm thickness as the result of the loss of a detoxification mechanism which has also been proposed as a role of LuxS (36). On balance, our observation supports an association between LuxS and quorum sensing in biofilm formation.
We attempted to complement the luxS mutation with respect to in vitro biofilm formation by the use of an AI-2 extract prepared from serovar Typhimurium. We did not, however, observe restoration of wild-type phenotype. Possibly, the Salmonella AI-2 could not bind to the appropriate L. reuteri receptor (whose identity is unknown), or the importance of LuxS is not in the production of autoinducer (and hence quorum sensing) but has some other regulatory significance, or biofilms may contain much higher localized concentrations than can be achieved exogenously in culture medium (4). The complemented strain of the luxS mutant was not considered appropriate for use in either in vitro or in vivo biofilm studies because the kinetics of AI-2 production were different from those of the wild type.
The ecological performance of mutant lactobacilli, as we have reported previously, is best tested in competition experiments (35). Lactobacillus johnsonii 100-5 and L. reuteri 100-93 were chosen as potential competitor strains because, according to the niche exclusion principle, bacterial strains with the same ecology cannot coexist in a community (8). Based on this reasoning, strain 100-5 and 100-23C would not be in competition because, belonging to different species, they would occupy different ecological niches. Strains 100-23C and 100-93, in contrast, both belonging to L. reuteri, would compete for the same niche. We did not use the wild-type strain as competitor because the AI-2 produced by this strain might have complemented (“cross-fed”) the mutant. So that the analytical culture method would be comparable (use of antibiotic resistance to measure the size of the mutant population relative to competitor), we used a xylA mutant of 100-23C for comparison of competitiveness. The xylA mutant, as reported previously, has the same ecological performance as the wild-type strain (35). The results of these experiments showed that the luxS mutant had reduced performance in the competitive conditions prevailing in the cecal ecosystem in the presence of a competitor of the same species. Although the reduced competitiveness of the mutant might have been due to the lack of autoinducer production, it could have been due to the decreased ability of the mutant strain to produce ATP via substrate level phosphorylation and/or to changes in SAM-related metabolic processes (9); effects that might only be manifested under the highly competitive conditions existing in the large bowel ecosystem. Strain 100-93 produced larger amounts of AI-2 compared to 100-5 in vitro, but this did not provide an ecological advantage over the xylA mutant. The amount of AI-2 produced in vitro might not be reflected in vivo but, if it was, it seems that sheer quantity of autoinducer alone does not deliver a competitive advantage.
Although we could not clarify the significance of AI-2 or AI-3 as autoinducers in Lactobacillus ecology, the results of our investigation clearly showed that inactivation of the luxS gene impacts heavily on the behavior of L. reuteri 100-23C when inhabiting the gastrointestinal tracts of mice. Of particular interest was the observation that the mutation had different consequences for the lactobacilli in different regions of the gut demonstrating niche-specific impacts of the luxS mutation: biofilm thickness on the forestomach epithelium increased but competitiveness in the large bowel was reduced. It should be possible to obtain mechanistic answers to the impact of LuxS on cellular processes in vitro and in vivo once the genome sequence of strain 100-23 becomes available (http:www.jgi.doe.gov/sequencing/why/CSP2005/lactobacillusreut.html). This will enable DNA microarrays representing the genome of 100-23 to be constructed and for in vitro and in vivo gene expression studies to be conducted.
Apart from demonstrating, for the first time, the importance of LuxS for the in vivo functionality of a gut commensal, our study has shown the utility of model systems (bioreactor, forestomach and cecum of Lactobacillus-free mice) that, together with the prospective use of DNA microarrays, provide a solid basis for future research concerning the importance of LuxS for bacterial life in the gut.
Acknowledgments
We are grateful for the support of the Institut Rosell-Lallemand, Inc., Montreal, Quebec, Canada, and of the Tangled Bank Research Cluster of the Department of Microbiology and Immunology, University of Otago.
We thank Vanessa Sperandio for AI-3 assays, Bonnie Bassler for V. harveyi MM32, and Sieu Tran for conducting the ATP assays. The assistance of the Otago Centre for Confocal Microscopy, Department of Anatomy and Structural Biology, Otago School of Medical Sciences, is also gratefully acknowledged.
REFERENCES
- 1.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blehert, D. S., R. J. Palmer, J. B. Xavier, J. S. Almeida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook, G. M., and J. B. Russell. 1994. Energy-spilling reaction of Streptococcus bovis and resistance of its membrane to proton conductance. Appl. Environ. Microbiol. 60:1942-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egland, P. G., R. J. Palmer, and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 101:16917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 6.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Delmuth. 2002. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller, R., and B. E. Brooker. 1974. Lactobacilli which attach to the crop epithelium of the fowl. Am. J. Clin. Nutr. 27:1305-1312. [DOI] [PubMed] [Google Scholar]
- 8.Hardin, G. 1960. The competitive exclusion principle. Science 131:1292-1297. [DOI] [PubMed] [Google Scholar]
- 9.Kaper, J. B., and V. Sperandio. 2005. Bacterial cell-to-cell signalling in the gastrointestinal tract. Infect. Immun. 73:3197-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lundin, A., and A. Thore. 1975. Comparison of methods for extraction of bacterial adenine nucleotides determined by firefly assay. Appl. Microbiol. 30:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyon, R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparoni. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 12.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markwell, M. A., S. M. Haas, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 14.McConnell, M. A., A. A. Mercer, and G. W. Tannock. 1991. Transfer of plasmid pAMβ1 between members of the normal microflora inhabiting the murine digestive tract and modification of the plasmid in a Lactobacillus reuteri host. Microb. Ecol. Health Dis. 4:343-355. [Google Scholar]
- 15.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signalling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 18.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171-179. [DOI] [PubMed] [Google Scholar]
- 19.Pirt, S. J. 1975. Principles of microbe and cell cultivation. Blackwell, Oxford, United Kingdom.
- 20.Redfield, R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10:365-370. [DOI] [PubMed] [Google Scholar]
- 21.Russell, W. M., and T. R. Klaenhammer. 2001. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl. Environ. Microbiol. 67:4361-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107-133. [DOI] [PubMed] [Google Scholar]
- 24.Savage, D. C., R. Dubos, and R. W. Schaedler. 1968. The gastrointestinal epithelium and its autochthonous bacterial flora. J. Exp. Med. 127:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 26.Schauder, S., K. Shokat, M. G. Surette, and B. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 27.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sperandio, V., A, G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sturme, M. H. J., M. Kleerebezem, J. Nakayama, A. D. L. Akkermans, E. E. Vaughan, and W. M. de Vos. 2002. Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Leewenhoek 81:233-243. [DOI] [PubMed] [Google Scholar]
- 30.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 32.Tannock, G. W. 1992. The lactic microflora of pigs, mice, and rats, p. 21-48. In B. J. B. Wood (ed.), The lactic acid bacteria in health and disease, vol. 1. Elsevier Applied Science, London, United Kingdom. [Google Scholar]
- 33.Tannock, G. W., C. Crichton, G. W. Welling, J. P. Koopman, and T. Midtvedt. 1988. Reconstitution of the gastrointestinal microflora of Lactobacillus-free mice. Appl. Environ. Microbiol. 54:2971-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter, J., N. C. K. Heng, W. P. Hammes, D. M. Loach, G. W. Tannock, and C. Hertel. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 69:2044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter, J., P. Chagnaud, G. W. Tannock, D. M. Loach, F. Dal Bello, H. F. Jenkinson, W. P. Hammes, and C. Hertel. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the gut of mice. Appl. Environ. Microbiol. 71:979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winzer, K., K. R. Hardie, and P. Williams. 2002. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 5:216-222. [DOI] [PubMed] [Google Scholar]
- 37.Winzer, K., Y.-H. Sun, A. Green, M. Delory, D. Blackely, K. R. Hardie, T. J. Baldwin, and C. Tang. 2002. Role of Neisseria meningitidis luxS in cell-to-cell signaling and bacteremic infection. Infect. Immun. 70:2245-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 39.Yuki, N., T. Shimazaki, A. Kushiro, K. Watanabe, K. Uchida, T. Yuyama, and M. Morotomi. 2000. Colonization of the stratified squamous epithelium of the nonsecreting area of horse stomach by lactobacilli. Appl. Environ. Microbiol. 66:5030-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]