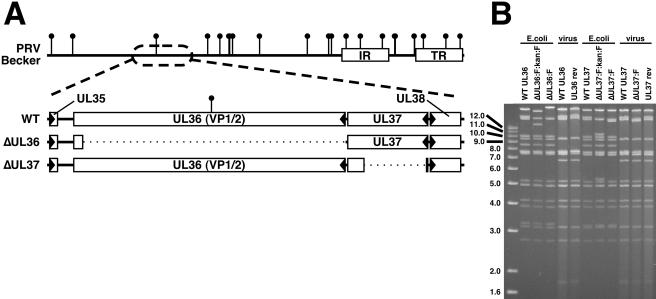
FIG. 1.
Description of VP1/2-null virus and VP1/2-complementing cell line. (A) Illustration of the PRV genome with characteristic internal and terminal repeats (IR and TR, respectively) shown as white rectangles. BamHI sites are indicated by vertical lollipops. The region of the genome relevant to this report is expanded to show the UL36 and UL37 ORFs, as well as the neighboring UL35 and UL38 ORFs (arrowheads indicate gene orientation). Below, dotted lines indicate regions deleted in the ΔUL36 (PRV-GS678) and ΔUL37 (PRV-GS993) viruses. (B) Ethidium bromide-stained agarose gel of BamHI-digested infectious clone DNAs isolated from E. coli and corresponding viral DNA isolated from nucleocapsids. The UL36 gene overlaps the two largest BamHI fragments of the viral genome, whereas the UL37 gene is exclusively on the second largest fragment. The FRT:kan:FRT insertion is ∼1.5 kbp and encodes two BamHI sites within the kanamycin resistance cassette, resulting in the truncation of the two large fragments during the ΔUL36 construction, and split of the second largest fragment into two smaller fragments during the ΔUL37 construction. Removal of the kanamycin resistance cassette and one FRT equivalent results in fusion of the remainders of the two largest fragments to produce the final ΔUL36 allele, and the reunion of the remainders of the second largest fragment to produce the final ΔUL37 allele. Additional fragment variations seen between the infectious clone plasmids and viral DNA result from loss of the E. coli vector backbone (as described in reference 36).