Abstract
The cationic molecule thrombin-induced platelet microbicidal protein 1 (tPMP-1) exerts potent activity against Staphylococcus aureus. We previously reported that a Tn551 S. aureus transposon mutant, ISP479R, and two bacteriophage back-transductants, TxA and TxB, exhibit reduced in vitro susceptibility to tPMP-1 (tPMP-1r) compared to the parental strain, ISP479C (V. Dhawan, M. R. Yeaman, A. L. Cheung, E. Kim, P. M. Sullam, and A. S. Bayer, Infect. Immun. 65:3293-3299, 1997). In the current study, the genetic basis for tPMP-1r in these mutants was identified. GenBank homology searches using sequence corresponding to chromosomal DNA flanking Tn551 mutant strains showed that the fourth gene in the staphylococcal mnh operon (mnhABCDEFG) was insertionally inactivated. This operon was previously reported to encode a Na+/H+ antiporter involved in pH tolerance and halotolerance. However, the capacity of ISP479R to grow at pH extremes and in high NaCl concentrations (1 to 3 M), coupled with its loss of transmembrane potential (ΔΨ) during postexponential growth, suggested that the mnh gene products are not functioning as a secondary (i.e., passive) Na+/H+ antiporter. Moreover, we identified protein homologies between mnhD and the nuo genes of Escherichia coli that encode components of a complex I NADH:ubiquinone oxidoreductase. Consistent with these data, exposures of tPMP-1-susceptible (tPMP-1s) parental strains (both clinical and laboratory derived) with either CCCP (a proton ionophore which collapses the proton motive force) or pieracidin A (a specific complex I enzyme inhibitor) significantly reduced tPMP-induced killing to levels seen in the tPMP-1r mutants. To reflect the energization of the gene products encoded by the mnh operon, we have renamed the locus sno (S. aureus nuo orthologue). These novel findings indicate that disruption of a complex I enzyme locus can confer reduced in vitro susceptibility to tPMP-1 in S. aureus.
Over the past decade, it has become increasingly clear that mammalian platelets play a critical role in innate host defense against the induction and progression of endovascular infections (9, 10, 25, 26, 47). This host defense property relates to the capacity of platelets to release a group of cationic antimicrobial peptides collectively known as platelet microbicidal proteins (PMPs) at sites of endovascular damage or infection (9, 19, 25, 26). Both rabbit and human platelets release functionally and structurally similar PMPs (43, 50). These peptides have potent microbiostatic and microbicidal activities in vitro against pathogens that commonly invade the bloodstream (i.e., Staphylococcus aureus, coagulase-negative staphylococci, viridans group streptococci, and Candida species) (44). Several PMPs contain functionally conserved structural themes: a C-terminal α-helical microbicidal domain and an N-terminal chemokine domain, linked by an interposing γ-core motif that is reiterated in diverse microbicidal chemokines (kinocidins) and classical cysteine-stabilized antimicrobial peptides (51, 52).
To date, the most extensively characterized PMPs are those released from platelets following stimulation by the procoagulant molecule thrombin (i.e., thrombin-induced PMPs [tPMPs]) (9, 10, 25, 43, 44, 50, 51). In humans with S. aureus bacteremia, we have previously shown that strains which are intrinsically susceptible to tPMPs (tPMPs) in vitro are less likely to cause complicated endovascular infections, such as endocarditis. In addition, such strains are more rapidly cleared from the bloodstream than strains exhibiting in vitro the tPMPr phenotype (12, 13). Similarly, S. aureus isolates from the nares of healthy humans are more susceptible to killing by tPMPs in vitro than strains from either the nares or bloodstream of hemodialysis patients, a population of bacteria experiencing extensive blood contact (V. G. Fowler, L. M. McIntyre, C. H. Cabell, L. B. Reller, G. R. Corey, T. Jones, K. Clark, M. R. Yeaman, and A. S. Bayer, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. E-004, p. 250, 2003).
Recent studies indicate that tPMPs target and permeabilize the S. aureus cytoplasmic membrane (CM), initiating the ensuing lethal mechanisms of these peptides. Intracellular targets affecting macromolecular synthesis and autolytic enzyme pathways have also been implicated in the staphylocidal mechanisms of tPMPs (45, 46, 48). The precise countermeasures by which S. aureus strains may resist the microbicidal activity of tPMPs remain incompletely defined but appear to include enhanced membrane fluidity (5), reduced transmembrane potential (ΔΨ) (21), and carriage of plasmids encoding transmembrane efflux pumps for cationic compounds (26).
We previously reported on a tPMPr S. aureus strain, ISP479R, a Tn551 transposon mutant of the tPMPs parental strain, ISP479C (10). A number of subsequent investigations in a model of experimental endocarditis have shown that, compared to its isogenic parental strain, ISP479R is cleared more slowly and to a lesser extent from target tissues, both during the natural history of the infection as well as during antimicrobial prophylaxis and therapy (9, 10, 25). The current study was designed to identify the genetic determinants corresponding to the site of the Tn551 insertion in strain ISP479R and correlate transposon disruption of this site with specific phenotypes that may contribute to reduced in vitro killing by tPMPs.
(This work was presented in part at the 103rd General Meeting of the American Society for Microbiology, May 2003, Washington, D.C., and at the 8th International Society for Cardiovascular Infectious Diseases Congress, October 2005, Charleston, S.C.).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains used in this study are listed in Table 1. S. aureus parental strain ISP479 (tPMP-1s) harbors plasmid pI258, which has a temperature-sensitive replicon and carries transposon Tn551, which confers erythromycin resistance. ISP479C is a well-characterized tPMP-1s S. aureus strain that is a spontaneously cured, plasmid- and transposon-free variant of ISP479 (10). We have previously detailed the construction of ISP479R by transposition of Tn551 (10). Strains ISP479C and ISP479R are virtually identical in the following phenotypic and genotypic parameters: colonial pigmentation, hemolytic characteristics, coagulase, DNase, pulsed-field gel electrophoretograms, biotyping, antibiograms, platelet adherence and aggregation capacities, growth kinetics, cell wall-associated protein A expression, and fibrinogen-binding capacity (10). Moreover, growth of ISP479R on Mueller-Hinton (MH) agar revealed no evidence of small colony variant formation. The broth macrodilution MICs for ISP479C and ISP479R of gentamicin were 0.25 and 0.5 μg ml−1 at final inocula of 5 × 105 and 5 × 106 CFU (cation-supplemented MH broth) (unpublished data).
TABLE 1.
Plasmids and bacterial strains
| Strain or plasmid | Genotype, phenotype, and/or description | Reference or source |
|---|---|---|
| E. coli DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 λ−gyrA96 relA1 | BRL |
| S. aureus strains | ||
| 6850 | Clinical isolate | 20 |
| ISP479 | pI258 Cadr, Penr Ars/Asir Tn511[ermC] | 37 |
| ISP479C | ISP479 cured of pI258, tPMP-1s | 10 |
| ISP479R | ISP479 snoD::Tn511[ermC] | 10 |
| ISP479R-complemented | ISP479R geh::pJM7989 (snoA-G), tPMP-1s | This study |
| ISP479R(pJM282) | ISP479R with pJM282 | This study |
| RN4220 | 8325-4, nitrosoguanidine-induced restriction mutant used as primary recipient for plasmids propagated in E. coli | 35 |
| SH1000 | 8325-4 rsbU positive, tPMP-1s | 16 |
| SH1000-98 | snoD::Tn551, tPMP-1r | This study |
| TxA | φ11/ISP479R × ISP479C,a tPMP-1r, snoD::Tn511[ermC] | This study |
| TxA complemented | TxA geh::pJM798 (snoA-G), tPMP-1s | This study |
| TxB | φ11/ISP479R × ISP479C, tPMP-1r, snoD::Tn551 | This study |
| TxB-1 compemented | TxB geh::pJM798 (snoA-G), tPMP-1s | This study |
| Plasmids | ||
| pCL55 | S. aureus integration-shuttle vector | 27 |
| pCRII | t-tail cloning vector | Invitogen Life Technologies, Carlsbad, CA |
| pIM36 | pBlueScript II KS(+)::tnp(Tn551) | 32 |
| pIM40 | pT7Blue::geh | 32 |
| pJM272 | pRN6680 with blaZ deleted | This study |
| pJM282 | pJM272 with a 1.85-kb EcoRI-ClaI fragment from pCL55 with φL54a int and attP, serves as a site-specific chromosomal integration vector for S. aureus | This study |
| pJM795 | pCRII with snoA-G flanked by NotI sites amplified from plasmid pNAS20 | This study |
| pJM798 | pJM272::snoA-G | This study |
| pNAS20 | pUC19 with mnhA-G (snoA-G) | |
| pRN6680 | pBluescript SKII(+) with a 2.9-kb DNA fragment with tetM | 34 |
| pT7Blue | t-tail cloning vector | Novagen, Madison, Wis. |
Phage/host × recipient.
We have also previously described the construction of two tPMPr transductants of ISP479R into the tPMPs parental strain, ISP479C, named TxA and TxB (3). ISP479 is of a genetic lineage related to S. aureus strain 8325-4, which contains a natural 11-bp deletion within the rsbU locus of the stress response operon, sigB (18). This deletion renders such strains partially sigB deficient in terms of stress responses (36). Since exposure of bacteria to cytoplasmic membrane-permeabilizing tPMPs is undoubtedly a substantial stress, perturbations in sigB function may potentially impact survival outcomes following peptide exposure. With this in mind, selected phenotypic and genotypic studies (detailed below) were performed in parallel with both ISP479-derived constructs as well as with transductants created using a rsbU-repaired variant (in which SigB activity is therefore restored to wild type) of ISP479 lineage, SH1000 (see construction details below) (16). As with tPMP-1r strains derived from ISP479C, SH1000 tPMP-1r derivatives grew as well as SH1000 in vitro.
In addition to the laboratory-derived strains described above, select studies included the clinical isolate S. aureus strain 6850. Strain 6850 was isolated from a patient with bacteremia, osteomyelitis, and soft tissue abscesses. This strain (which is tPMP-1s) has been used in our laboratories in studies of antimicrobial peptides and transmembrane potential in S. aureus (20). This strain was principally used in the proton motive force (PMF) inhibitor investigations (see below).
For routine growth, S. aureus strains were cultivated in either brain heart infusion (BHI) broth (Difco, Detroit, MI) or tryptic soy broth (Difco Laboratories, Detroit, MI) with rotary agitation (250 rpm) or on medium augmented with 1.5% agar at 37°C. Escherichia coli strains were grown in Luria-Bertani (LB) broth with rotary agitation or on LB agar plates incubated at 37°C. For S. aureus, tetracycline and erythromycin selection used medium supplemented with antibiotic at a concentration of 10 μg and 5 μg ml−1, respectively. Resistant E. coli was grown in medium containing 100 μg ml−1 ampicillin or 12.5 μg ml−1 tetracycline. All strains were stored at −70°C.
Genotypic studies: genetic linkage analysis and construction of tPMP-1r variants in SH1000.
To confirm a genetic linkage between Tn551 in ISP479R and the in vitro tPMPr phenotype, cotransduction studies were performed as previously described using phage φ11 lysates from strain ISP479R (11). Generalized transduction of the rsbU-repaired, tPMPs parental strain, SH1000, was performed at a multiplicity of infection of a 1:10 ratio of bacteriophage to recipient cells. Transductants were selected on 0.3% glycerol-lactate agar containing erythromycin, and tPMPr candidates were initially identified by resistance to the cationic antimicrobial peptide protamine (used in a number of previous studies as a screening surrogate antimicrobial peptide in place of tPMPs [10]). Candidate transductants were then checked for a stable tPMPr phenotype using a standard microtiter killing assay as previously described (5, 44). S. aureus transductant SH1000-98 was identified by this protocol and used in the subsequent studies described below.
Southern hybridization.
We have previously shown that ISP479C is Tn551 free, while in strains ISP479R and TxB, the transposon resides within similarly sized restriction endonuclease fragments (10). To verify the genotype of the strains used in this study and to provide additional information on the size of the Tn551-containing DNA fragment for estimating the appropriate amplification times for inverse PCR, we probed the chromosome of the strains used in this study for Tn551 by Southern hybridization.
Briefly, staphylococcal chromosomal DNA was isolated (11) and subjected to electrophoresis through 0.7% agarose gels. DNA was transferred to nylon membranes (MagnaGraph; Fisher Scientific, Pittsburgh, PA) and probed using either digoxigenin- or [α-32P]dCTP-labeled probes (as noted in the figure legends). The probes used for Southern analysis included a ≈1.2-kb HindIII fragment encoding the Tn551 transposase from plasmid pIM36 and a ≈2.7-kb BamHI-HindIII fragment encoding the gene for glycerol ester hydrolase (geh) from plasmid pIM40. Digoxigenin-labeled probes were prepared as described for use with the Genius system (Boehringer Mannheim, Indianapolis, IN). Prehybridization and hybridization used standard buffer plus 50% formamide according to the manufacturer's instructions. Chemiluminescent probe detection used the substrate disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.2.237]decan}-4-yl)phenyl phosphate (Boehringer Mannheim). Radiolabeled probes were prepared using the Prime-a-Gene labeling kit (Promega, Madison, WI) according to the manufacturer's instructions. Prehybridization and hybridization used 1× SSC (0.15 M NaCl and 0.015 M sodium citrate), 1% sodium dodecyl sulfate (SDS), 0.5% formamide, Denhardt's solution (0.04% Ficoll 400, 0.04% polyvinylpyrrolidone, and 0.04% bovine serum albumin, fraction V), and 2.5 mg ml−1 yeast RNA. All membranes were prehybridized for 4 h and hybridized for 12 h at 42°C, washed twice with 2× SSC containing 0.1% SDS for 5 and 15 min at room temperature, and washed once each in 0.5× SSC, 0.1% SDS for 15 min, 0.1× SSC, 0.1% SDS for 15 min, and 0.1× SSC, 1.0% SDS for 30 min. The later washes were performed at 68°C. Probes were detected by exposure of Kodak X-Omat AR film.
Identification of the transposon insertion site in tPMPr strains.
To identify the Tn551-disrupted gene in tPMPr strains ISP479R, TxB, and SH1000-98, DNA flanking the site of the transposon insertion site was amplified by inverse PCR and sequenced, with the sequence of the amplified product then used to search GenBank. Chromosomal DNA isolated from the mutant strains was prepared by digestion with PstI. The digested DNA was self-ligated, purified by phenol extraction, concentrated by ethanol precipitation, and used as the template for PCR. PCRs consisted of 0.5 μg DNA, 1 U high-fidelity polymerase mix (Boehringer Mannheim, Indianapolis, IN), 200 μM nucleotides, and 1.0 μM primers in each of seven 100-μl reaction mixtures that contained a range of 0.0 to 8.5 mM MgCl2 (8). Primers were designed to extend outward from the 5′ and 3′ ends of Tn551 (GenBank accession no. Y13600): 5′-GCGGCCGCTCATTAATGACACGGTGGAATG and 5-5GCGGCCGCGTTTCGTGTCGGGTCAATGATTG, respectively.
Amplified products were analyzed by agarose gel electrophoresis and by nucleotide sequence determination. Sequence data were obtained using an Applied Biosystems 377 DNA sequencer with dye terminator cycle sequencing chemistry (Perkin-Elmer, Foster City, CA) on a QIAquick PCR purification kit (QIAGEN, Chatsworth, CA), with purified templates initially using the Tn551 primers and, later, primers designed to the determined sequence. Sequence was compiled and analyzed using Sequencher version 4.1 (Gene Codes, Ann Arbor, MI) and Blastx (1), respectively.
Second-site complementation.
As noted in the Results section below, the transposon disruption mapped to the mnh operon (mnhABCDEFG) of S. aureus (15). For complementation studies, the wild-type mnh operon (from plasmid pNAS20; kindly provided by T. Hiramatsu, Tsushima, Okayama, Japan) was integrated into the chromosomal gene encoding lipase in S. aureus strains ISP479R, TxA, and TxB, using well-established techniques (27). The integration vector, pJM798, was constructed from plasmid pRN6680. Plasmid pRN668 is the cloning vector pBluescript (Stratagene, La Jolla, CA) with a 2.9-kb DNA fragment encoding a staphylococcal tetracycline resistance marker, tetM (25). The gene encoding β-lactamase was deleted from pRN6680 by BglI digestion followed by intermolecular ligation creating pJM272. A 1.85-kb EcoRI-ClaI fragment from plasmid pCL55 (27), containing the L54a att site, and integrase was cloned into the similar site in pJM272, creating plasmid pJM282. The mnh operon was amplified from plasmid pNAS20 t-tail cloned into pCRII, creating pJM795 (15). The mnh-specific primers with 5′-appended NotI sites were 5′-GCGGCCGCTCATTAATGACACGGTGGAATG and 5′-GCGGCCGCGTTTCGTGTGGGTCAATGATTG. Plasmid pJM795 was digested with NotI, and the mnh-containing fragment was transferred into a similar site within pJM282, creating plasmid pJM798 (see Fig. 5, below). Plasmid pJM798 was introduced into S. aureus strain RN4220 by electroporation and then transduced to ISP479R, TxA, and TxB using bacteriophage φ11, selecting for pJM798-encoded tetracycline resistance using previously described procedures (2, 32). Correct chromosomal insertion was verified by Southern analysis.
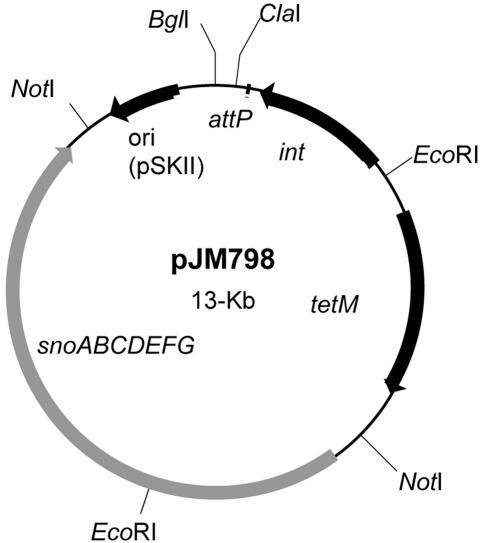
FIG. 5.
Plasmid map of the integrative complementation plasmid, pJM798, showing relevant restriction sites and genes. The sno (mnh) operon, E. coli col E1 origin of replication (ori) (pSKII), bacteriophage L54a attachment site (attP), integrase gene (int), and the tetracycline resistance marker (tetM) are delineated by arrows or boxes.
As a control for the impact of lipase locus disruption on tPMP-1 susceptibility phenotypes, we constructed strain ISP479R(pJM282), which is ISP479R carrying a chromosomal integration of pJM282 (without mnhABCDEFG). The integration vector was introduced into ISP479R by transduction of tetracycline resistance encoded on a chromosomal copy of pJM282 in PM291.
Phenotypic studies: preparation of tPMP-1.
For this study, we focused on tPMP-1 since it is the most abundant of the tPMPs in rabbit platelets (a standard in vivo model of S. aureus endovascular pathogenesis [25, 50]). Supernatants from thrombin-stimulated rabbit platelets were prepared as previously described (50). Purification of tPMP-1 was achieved by column chromatography followed by reversed-phase high-performance liquid chromatography. Bioactivity of tPMP-1 preparations was validated as previously published (43, 50). Purified and bioactive tPMP-1 was lyophilized and kept frozen at −20°C until thawed for use.
Standard tPMP-1 susceptibility assay.
Susceptibility profiles of the study constructs to tPMP-1 were determined by timed-kill assay as described elsewhere (44). Overnight cultures of the S. aureus isolates were washed twice and suspended in phosphate-buffered saline. Dilutions of tPMP-1 and S. aureus isolates were added to polypropylene microculture tubes containing Eagle's minimal essential medium (Irvine Scientific, Santa Ana, CA) to achieve a final tPMP-1 concentration of 1 or 2 μg ml−1 and a final bacterial inoculum of 103 CFU ml−1 (e.g., the standard inoculum and peptide concentration used for tests of this peptide against S. aureus strains [43]). For determinations of percent survival, one tube contained only bacteria and minimal essential medium and acted as a positive growth control. After 2 h of incubation at 37°C, suspensions were mixed by vortexing, and 15-μl aliquots were removed from each tube and quantitatively cultured onto blood agar plates. The proportion of S. aureus cells of each construct surviving a 2-h exposure to tPMP-1 was expressed as the percent CFU of the positive growth control. All assays were performed in triplicate on two separate days, and the mean percent survival ± the standard deviation (SD) was determined. A tPMP-1r phenotype was defined as ≥50% survival of the initial inoculum after a 2-h exposure to 1 or 2 μg ml−1 tPMP-1 at 37°C. This arbitrary in vitro breakpoint correlates with an enhanced capacity of such strains in vivo to induce and propagate human or experimental infective endocarditis (12, 25, 44).
Standard protamine susceptibility assay.
All study strains were also tested for their MICs to protamine by an agar dilution assay. As noted above, protamine is a cationic antimicrobial peptide from salmon sperm whose in vitro activity against S. aureus tends to closely parallel that of tPMP-1 (9, 10). Prior studies have shown the MICs of strains ISP479C and ISP479R to be 1 to 2 versus 8 mg ml−1, respectively (10). Briefly, protamine sulfate (Sigma Chemical, St. Louis, MO) was incorporated into LB agar plates over a range of concentrations from 1 to 16 mg ml−1. After inoculating 5 × 107 CFU onto their surface, the plates were incubated for 48 h at 37°C. The MIC was determined as the lowest protamine concentration inhibiting all visible growth.
Membrane fluidity assays.
Increased membrane fluidity has been a consistent feature of most tPMP-1r S. aureus isolates analyzed to date (5). Thus, we assessed this parameter in our study strains by quantifying the relative distribution and two-dimensional orientation of a fluorescent dye within the lipid layers of the cytoplasmic membrane. In brief, a whole-cell suspension of each bacterial strain was prepared with a bacterial density of ∼109 CFU ml−1. Cells were then labeled with a fluorescent dye, 1,6-diphenyl-1,3,5-hexatriane (DPH; Sigma Chemical, St. Louis, MO) (5). DPH specifically labels and fluoresces within the hydrophobic regions of the lipid bilayer but lacks fluorescence in aqueous environments (5). The steady-state fluorescence of the DPH probe predominantly reflects the structural order of membrane lipids, as a result of partitioning of the probe into the hydrocarbon phase of the lipid membrane bilayer when it is in a fluid state. Thus, membrane fluidity may be considered as the reciprocal of the structured order of membrane lipids. For DPH labeling, a 2 mM solution of this dye was prepared in tetrahydrofuran, and 100 μl was added to 50 ml of agitated 0.05 M Tris-HCl (pH 7.6). Excess tetrahydrofuran was removed by flushing with nitrogen. Cells were then incubated in DPH at a final concentration of 2 μM for 3 h at 30°C.
Fluorescence polarization was measured using a Kontron spectrofluorometer (Copenhagen, Denmark) as previously detailed (5). Excitation of the fluorescent probe was accomplished with vertically polarized monochromatic light at 360 nm for DPH, with emission intensity quantified at 426 nm, using a detector oriented either parallel to or perpendicular to the direction of the polarized excitation source. The degree of fluorescence polarization or polarization index (PI) was calculated as described previously (5). The lower the PI value, the more fluid the membrane.
Membrane potential (ΔΨ) assays.
For the determination of ΔΨ, 1 μCi of radiolabeled tetraphenylphosphoniumbromide ([3H]TPP+; Amersham, Piscataway, NJ) corresponding to 0.04 nM TPP+ was added to 1-ml culture aliquots of 108 CFU of bacteria. This lipophilic cation diffuses across the bacterial membrane in response to the membrane potential, with the negative pole being inside the cells. Cells were grown in chemically defined medium as previously reported (4, 42) at 37°C with agitation, and aliquots were removed serially over a 6-h period for determination of the optical density at 600 nm (OD600) and for ΔΨ measurements. For the latter analyses, samples of 100 μl were filtered on cellulose acetate filters (pore size, 0.2 μm; Schleicher & Schuell, Keene, NH) and washed twice with 5 ml 50 mM potassium phosphate buffer. Filters were dried, transferred to counting vials filled with scintillation fluid (Quickzint 100; Zinsser Analytic, Frankfurt, Germany), and counted in a Packard liquid scintillation analyzer (Tri-Carb 1900 CA). Counts were corrected for nonspecific binding of TPP+ to bacterial cells by subtracting the radioactivity retained by cells after treatment with 10% butanol. The membrane potential was calculated by means of the Nernst equation, ΔΨ = (2.3 × R × T/F) × log([TPP+in]/[TPP+out]), with R being the universal gas constant, T the absolute temperature in Kelvin, F the Faraday's constant, [TPP+in] the molar concentration of TPP+ inside the bacterial cells, and [TPP+out] the molar concentration of TPP+ in the medium. The internal volume of 3.4 μl mg−1 protein of staphylococcal cells used for calculation of [TPP+in] was determined previously (4, 42).
Staphylococcal growth in hypertonic saline and at various pHs.
Halotolerance of the study strains was tested using standard BHI broth or BHI broth supplemented with NaCl at a final concentration range of 50 mM to 3 M (pH 7.2), with a starting inoculum of ∼104 CFU ml−1 of bacteria. In parallel, pH tolerance was determined using similarly inoculated BHI broth, adjusted with either HCl or NaOH to provide a final pH range from 4.5 to 8.5. All cultures were incubated at 37°C with rotary agitation. For halotolerance, the OD600 was measured in culture aliquots obtained at even-hour intervals for 10 h. For pH tolerance, OD600 measurements and comparisons were made on samples obtained at 8 h (41).
PMF and complex 1 inhibitor studies.
To decrease the PMF, we used carbonylcyanide m-chlorophenylhydrazone (CCCP; Sigma Chemical), an uncoupler of oxidative phosphorylation. Test strain ISP479C and the clinical isolate 6850 were grown to stationary phase at 37°C in MH broth, because this medium contains very little glucose compared to brain heart infusion broth (high glucose concentrations reverse the effects of CCCP [33]). Cells were harvested, washed twice in buffered saline, added to fresh MH broth, and grown for 30 min at 37°C in the presence or absence of 100 μM CCCP. Cells were then harvested and processed through the standard tPMP-1 susceptibility assay described above, using a final peptide concentration range of 0.25 to 2 μg−1. Piericidin A is a specific inhibitor of complex I NADH oxidoreductases (6). For piericidin A studies, the same protocol as used for CCCP was followed, only substituting piericidin A (4.0 or 8.0 μM) for CCCP.
Statistical analysis.
Continuous data were analyzed by the Kruskal Wallis analysis of variance, with the Tukey post hoc correction for multiple comparisons where appropriate. A P value of ≤0.05 was considered significant.
RESULTS
Southern analysis of isogenic constructs.
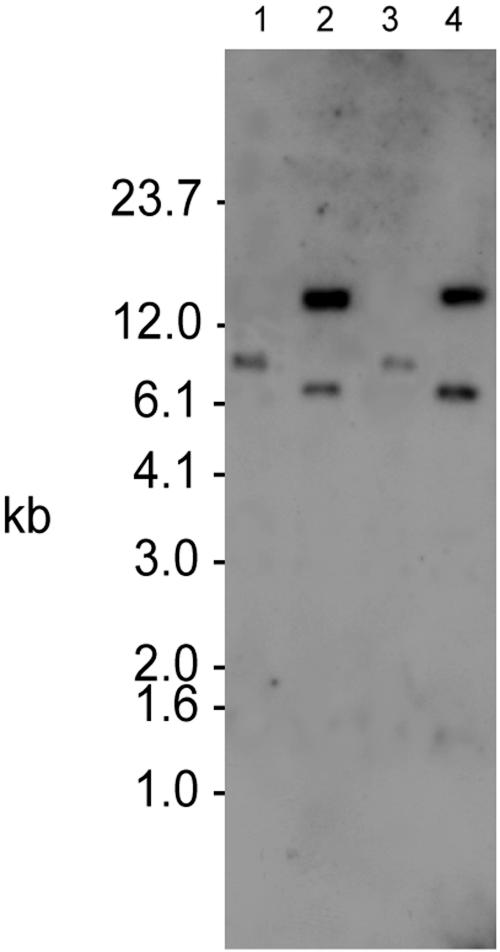
The Tn551 probe detected a single DNA fragment in ISP479R that was not in the parental strain (Fig. 1). Chromosomal DNA from ISP479R digested with EcoRI resulted in identification of a 13-kb fragment, while the probe hybridized with a 10.2-kb PstI-digested DNA fragment. These insert sites are identical to those previously identified and reported (10). Also, as previously reported, Southern analyses confirmed the presence of a single Tn551 insert in the tPMP-1r transductant constructs, TxA and TxB (data not shown) (10). Within the tPMP-1r strain SH1000-98, Tn551 resided on restriction fragments identical in size to those associated with ISP479R (Fig. 2).
FIG. 1.
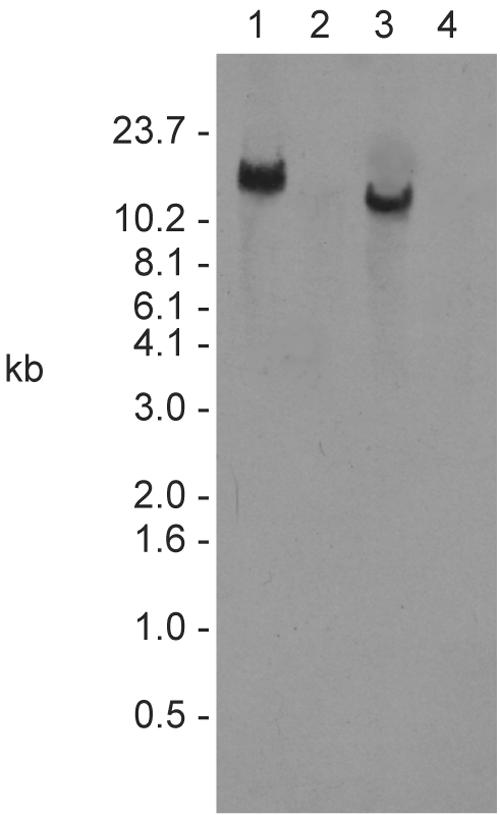
Southern analysis of DNA isolated from the tPMP-1r transposon mutant ISP479R (lanes 1 and 3) and parental tPMP-1s, ISP479C (lanes 2 and 4), probed with a transposon-specific probe. Chromosomal DNA was digested with EcoRI (lanes 1 and 2) or PstI (lanes 3 and 4). A probe that hybridizes with the gene encoding the transposase of Tn551 detected a single 13-kb EcoRI fragment and a 10.2-kb PstI fragment in strain ISP479R that was not in parent strain ISP479C.
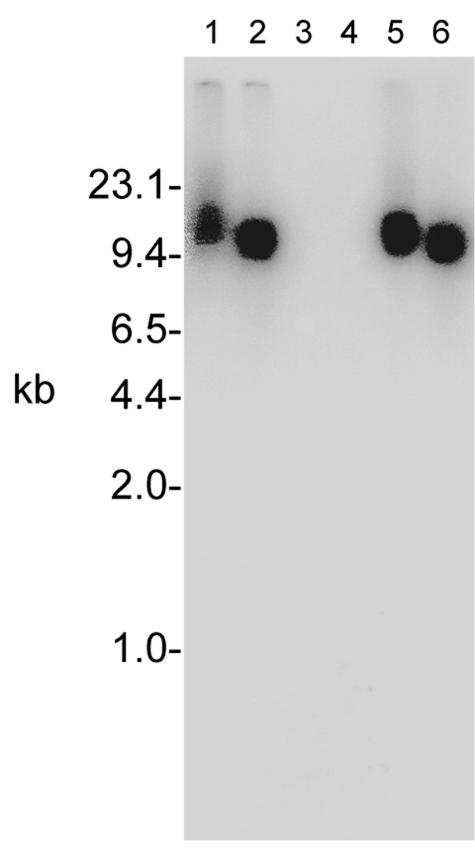
FIG. 2.
Southern analysis of S. aureus SH1000-98, a PMP-1r transductant derived from ISP479R. Lanes 1 and 2 contain chromosomal DNA from ISP479R, lanes 3 and 4 contain chromosomal DNA from strain SH1000, and lanes 5 and 6 contain DNA from SH1000-98. The even-numbered lanes contain PstI-digested DNA, while the odd-numbered lanes contain EcoRI-digested DNA. A Tn551-specific probe detected similarly sized DNA fragments in ISP479R and SH1000-98 that were not present in SH1000 or ISP479C (Fig. 1).
Identification of the transposon insertion site.
Inverse PCR using Tn551-derived primers on PstI-digested chromosomal DNA from strains ISP479R, TxA, TxB, and SH1000-98 amplified a single product. The concentration of MgCl2 used in the amplification reaction mixtures affected formation of the product. For each strain, no specific DNA amplification was observed when less than 3.0 mM MgCl2 was used in the reaction mixture, while concentrations of MgCl2 that ranged from 3.0 to 9.0 mM resulted in amplification of a 5-kb DNA fragment (Fig. 3, ISP479R). The size of this fragment is consistent with the Southern hybridization data. The 10.2-kb PstI-hybridizing fragment corresponds to the size of the amplified product plus the 5.2 kb of DNA comprising Tn551.
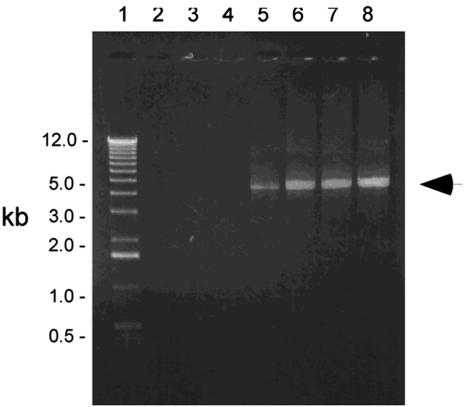
FIG. 3.
Amplification of the Tn551 insert site in strain ISP479R. A 0.75% agarose gel with the 1-kb ladder (lane 1) and five independent inverse PCRs that used a PstI-generated template and MgCl2 concentrations ranging from 0.0 to 9 mM in 1.5 mM increments (lanes 2 to 5). An arrow marks a 5-kb product. The size of this fragment is consistent with the Southern blot data (Fig. 1); the 10.2-kb PstI fragment corresponds to the size of the amplified product plus the 5.2 kb of DNA comprising Tn551. Similar DNA band patterns were observed for TxA, TxB, and SH1000-98 (data not shown).
Nucleotide sequences of 540 bp and 65 bp were directly determined from the PCR products, which corresponded to DNA flanking the 5′ and 3′ ends of the transposon, respectively. Using these data as the query sequence in Blastx searches of published S. aureus genomes, we found that the transposon had inserted into the gene previously identified as mnhD. The mnhD locus encodes a putative subunit of a Na+/H+ antiporter encoded by mnhABCDEFG (15). The quality of the data generated by sequencing the sense and antisense DNA from the above tPMP-1r constructs allowed for accurate mapping of the transposon insertion site to within a 65-bp segment of chromosome and for determination of the orientation of the transposon relative to the disrupted gene (Fig. 4).
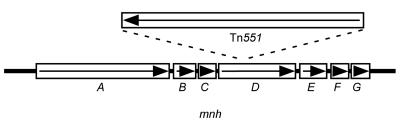
FIG. 4.
Map of the Tn551 insertion site in strains ISP479R, TxA, TxB, and SH1000-98. The sno (mnh) operon is shown and consists of six genes (rectangles labeled A through G). Tn551 is depicted by the rectangle above the map of the sno (mnh) operon. The arrows within the depicted genes indicate the orientation of transcription of snoA-G (mnhA-G) and Tn551. The converging lines from the ends of the representation of Tn551 show the approximate location of the transposon insertion site.
Sequence searches in GenBank revealed that, in addition to a putative Na+/H+ antiporter, the gene containing the transposon insertion site encodes a protein with homology to members of the NADH-ubiquinone/plastoquinone (complex I) enzyme family (pfam 0036; expectation = 6e−22). Included in this protein family are the E. coli complex I-coding subunits encoded by nuoL, nuoM, and nuoN (expectation = 7e−21, 1e−26, and 5e−24, respectively) (29). GenBank searches also identified similar proteins in a variety of organisms. Outside of the database entries for staphylococcal species, the gene with the highest homology to the wild-type allele of the disrupted gene was mpr (expectation = e−110), a Bacillus subtilis complex I-like-encoding operon that reportedly confers Na+ and cholate resistance and maintains pH homeostasis (17).
Based on the above genotypic analyses and the phenotypic studies detailed below, we renamed the disrupted operon snoABCDEFG (snoA-G, for staphylococcal nuo-like orthologues A through G). This nomenclature will be used for the remainder of the paper.
Second-site complementation of snoD.
It is conceivable that snoD may be the only gene within the snoA-G operon whose function is adversely affected by the Tn551 mutation. However, it is also possible that, depending on transcriptional content and translational start and stop sequences of the sno messages, additional genes upstream or downstream of snoD within the operon may have altered expression. Given this possibility, we utilized the entire seven-gene sno operon for second-site complementation studies.
A wild-type copy of the operon, carried on plasmid pJM798 (Fig. 5), was integrated into the chromosomal gene encoding glycerol ester hydrolase (lipase gene, geh) in S. aureus strains ISP479R and TxB. Interruption of the chromosomal copy of this gene by integration of plasmid pJM798 was examined by Southern analysis in strains ISP479R, ISP479R with pJM798, TxB, and TxB with pJM798 (Fig. 6). The lipase gene resides within an 8-kb fragment in strains without pJM798 integrated into the chromosome. Upon plasmid integration into the lipase gene, an EcoRV site within pJM798 results in two fragments. The sizes of the two fragments correspond to the sum of the geh EcoRV fragment in the wild-type strain plus the 13 kb of pJM798. These studies confirmed the successful integration of pJM798 into the targeted chromosomal locus in the tPMP-1r constructs.
FIG. 6.
Southern analysis of plasmid pJM798 second-site-complemented strains. EcoRV-digested chromosomal DNA from ISP479R (lane 1), ISP479R [chr::pJM798] (lane 2), TxB (lane 3), and TxB [chr::pJM798] (lane 4) was probed with the gene encoding staphylococcal glycerol ester hydrolase (geh). Upon integration into the chromosome, plasmid pJM798 introduces an EcoRV site that results in two fragments with a combined size equal to the sum of the geh EcoRV fragment in the parental strain plus the 13 kb of pJM798.
Antimicrobial peptide susceptibility profiles.
As shown in Table 2, compared to their parental strain (ISP479C), the original Tn551 mutant (strain ISP479R) and transductants derived from ISP479R (strains TxA and TxB) exhibited reduced susceptibility to tPMP-1 and protamine. With regard to strains with the ISP479C genetic background, these data are consistent with our previously reported results (10). Of note, the reduced susceptibility to tPMP-1 for the transductant strains was virtually identical to the original transposon mutant (ISP479R). S. aureus strain SH1000-98, a transductant derived from ISP479R that carries the defined Tn551 mutation, exhibited reduced susceptibility to tPMP-1 compared to its parent strain, SH1000. Second-site complementation with a wild-type copy of the sno operon in the original transposon mutant, as well as both TxA and TxB, restored a tPMP-1s phenotype, although the susceptibility profile did not fully reach parental levels. The ISP479R-derived strain in which the lipase gene was disrupted by the plasmid vector alone (without the snoA-G complementation) remained tPMP-1r.
TABLE 2.
tPMP-1 and protamine susceptibility profiles
| Strain | % Survival
|
MIC of protamine (mg ml−1)b | Profilec | |
|---|---|---|---|---|
| 1 μg ml−1 tPMP-1a | 2 μg ml−1 tPMP-1a | |||
| ISP479C | 27 ± 1.4*d | 20 ± 2.1* | 1 | S |
| ISP479R | 84 ± 2.1 | 75 ± 3.5 | 8 | R |
| ISP479R-comp | 44 ± 2.8 | 35 ± 2.1 | 1 | S |
| ISP479R(pJM282) | 93 ± 6.0 | 95 ± 8.0 | NDe | R |
| TxB | 84 ± 6.4 | 82 ± 3.5 | 8 | R |
| TxB-comp | 47 ± 2.8 | 47 ± 8.5 | 1 | S |
| SH1000 | 31 ± 5.0 | 21 ± 10.0* | 1 | S |
| SH1000-98 | 66 ± 6.0 | 59 ± 2.0 | 8 | R |
Mean ± SD of three independent runs.
MIC range on three independent runs.
sensitive (S) and resistant (R) to tPMP-1 and protamine as defined in Materials and Methods.
P < 0.05 comparing the parental strain to its respective tPMP-1r mutant.
ND, not determined.
All tPMP-1r strains also exhibited an eightfold increase in their MICs of a second cationic bactericidal peptide, protamine, compared to their respective parental strains. Of note, second-site complementation of all the tPMP-1r strains restored parental-level protamine MICs in these constructs.
Cytoplasmic membrane fluidity.
We investigated the impact of the snoD disruption, as well as sno operon second-site complementation, upon CM fluidity by fluorescence polarization spectrofluorometry. As shown in Table 3, CM fluidity was significantly higher (i.e., lower PI values) for tPMP-1r strains ISP479R and SH1000-98 than for their respective parental strains (P < 0.05 for all comparisons of parent versus mutant). Similar data were observed for TxB (not shown). Furthermore, complementation with the sno operon in ISP479R and TxB reversed the altered CM fluidity to a more rigid state (not shown).
TABLE 3.
Comparative membrane fluidity profiles
| Strain | PI (± SD)a | P value |
|---|---|---|
| ISP479C | 0.320 ± 0.020 | <0.05 vs ISP479C and ISP479R-comp |
| ISP479R | 0.308 ± 0.010 | |
| ISP479R-comp | 0.320 ± 0.010 | |
| SH1000 | 0.325 ± 0.025 | <0.05 vs SH1000 |
| SH1000-98 | 0.301 ± 0.025 |
See Materials and Methods for methodologic details.
Membrane potential assays.
Baseline ΔΨ values of the parental, transposon mutant, and transductant mutant in the ISP479C background were similar and consistent with data from other tPMP-1s parental strains (20) (Table 4). Although baseline ΔΨ fell modestly during late-logarithmic growth, the parental strain reestablished a near-parental ΔΨ during early postexponential growth. In contrast, neither tPMP-1r construct could reestablish parental-level ΔΨ over this same growth period.
TABLE 4.
Changes in membrane potential during growth
| Time of measurement (h) | Change in Δ ψ (mV)
|
||
|---|---|---|---|
| ISP479C | ISP479R | TxB | |
| 0 | 123 | 129 | 144 |
| 0.5 | 119 | 96 | 123 |
| 1 | 113 | 83 | NDa |
| 2 | 114 | 91 | 98 |
| 3 | 103 | 93 | 72 |
| 4 | 107 | 73 | 74 |
| 6 | 116 | 82 | 100 |
ND, not done.
Halotolerance and pH tolerance.
Previous investigators proposed that the mnh (sno) operon encodes a multisubunit antiporter in S. aureus that only transports Na+/H+ ions in response to their distribution relative to the bacterial membrane and, thus, is a secondary antiporter (15). A secondary antiporter, but not a complex I NADH oxidoreductase, would be expected to substantially impact halotolerance and pH tolerance of the organism (15). Moreover, complex I enzymes can be distinguished from enzymes involved in later stages of the electron transport system (e.g., complex III enzymes) by their effects upon growth at alkaline pH (pH ≥ 8.5) (41). Therefore, we assessed the impact of the transposon mutation in ISP479R and SH1000-98 on putative antiport function by comparing the growth kinetics of these strains to their parental controls in media containing various NaCl concentrations or buffered to different pH levels.
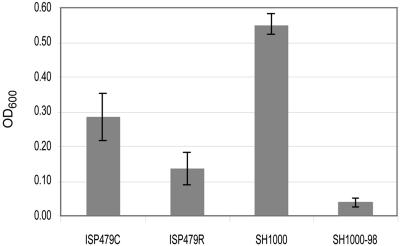
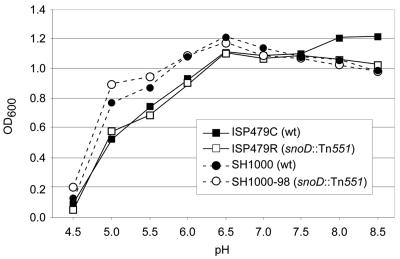
All strains grew equally well at a range of NaCl concentrations of 50 mM to 1.5 M (data not shown). At NaCl concentrations of 3 M, all strains experienced a relatively long lag phase (∼8 h) before logarithmic growth ensued (data not shown). Both tPMP-1s parental strain constructs grew better than their mutant derivatives between 8 and 10 h of incubation (Fig. 7). Furthermore, within a pH range between 4.5 and 8.5, the relative growth profiles of all four strains were similar over a 24-h incubation period (Fig. 8). Of interest, growth of SH1000 generally exceeded that of ISP479C in both high NaCl concentrations and under low-pH conditions, probably reflecting the intact sigB stress operon in the former versus the latter strain (16).
FIG. 7.
Halotolerance of parental versus respective PMP-1r mutants grown for 10 h at 37°C in NaCl-supplemented medium (3 M NaCl). Data represent the means (±SD) of three independent experiments.
FIG. 8.
pH tolerance of parental versus respective PMP-1r mutants grown for 24 h at 37°C in medium adjusted to a range of pHs from 4.5 to 8.5. Data represent the mean OD600 values obtained at 24 h growth for three independent experiments. SD bars are omitted for clarity of figure presentation.
PMF inhibitor studies.
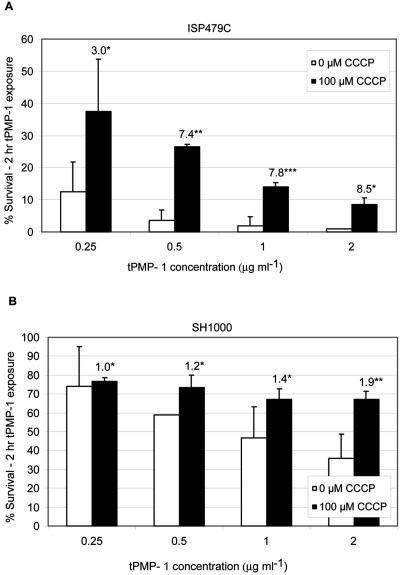
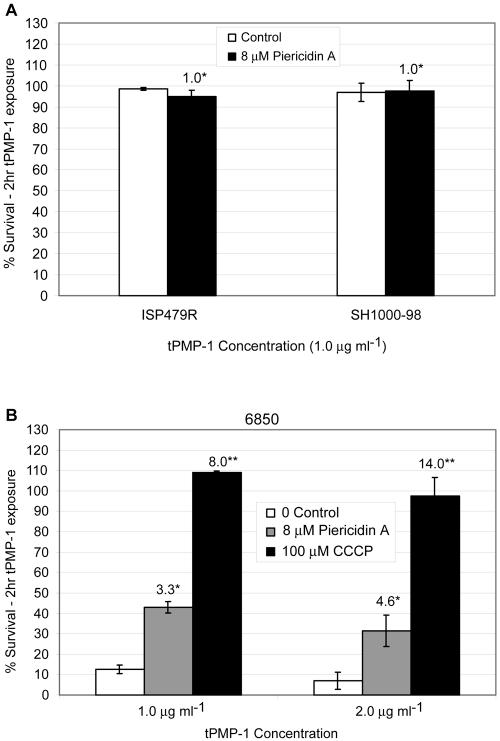
Unlike Na+/H+ antiporters, complex I NADH oxidoreductases function to generate a proton gradient and establish the PMF. However, these enzymes can exhibit a secondary Na+ antiport activity (6). Conversely, secondary Na+/H+ antiporters minimally contribute to generation of a proton gradient and, thus, have no effect on establishing the PMF. The PMF inhibitor CCCP caused significantly reduced killing of both tPMP-1S parental strains ISP479C and SH1000 over a range of tPMP-1 concentrations (Fig. 9). CCCP impacted tPMP-1 susceptibility in a manner that was substantially greater in ISP479C than in SH1000 (3- to 8.5-fold versus a ∼2-fold increase in survival in the presence of tPMP-1, respectively). As anticipated, CCCP did not impact tPMP-1 susceptibility profiles in tPMP-1r strains (data not shown).
FIG. 9.
Impact of CCCP treatment of tPMP-1s parental strains upon baseline tPMP-1 susceptibility profiles. Data represent the mean (±SD) of surviving CFU after exposure to various concentrations of tPMP-1 (0 to 2 μg/ml) in the presence or absence of CCCP (100 μM). (A) Data for parental strain ISP479C. *, not significantly different from untreated controls; **, P < 0.0001 versus untreated controls; ***, P < 0.001 versus untreated controls. (B) Data for parental strain SH1000. *, not significantly different from untreated controls; **, P < 0.05 versus untreated controls. Numbers above bars in panels A and B represent the mean fold increases in surviving CFU, comparing CCCP exposures to no-CCCP exposures. These data represent the means of at least two independent assays.
Complex I enzyme inhibitor studies.
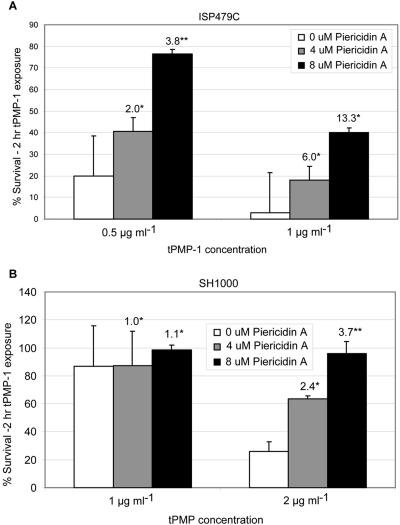
To test the prediction that disruption of a complex I enzyme results in a more tPMP-1r phenotype, parental strains ISP479C and SH1000 were grown in the presence of the specific complex I inhibitor, piericidin A. Similar to the impact of CCCP above, in the presence of 4 or 8 μM piericidin A, there was a significant increase in survival of both tPMP-1s parental strains in tPMP-1 compared to cells grown in the absence of this inhibitor (Fig. 10). As seen with CCCP, the extent of the piericidin A effect was greater in strain ISP479C than SH1000. Similar to data from CCCP exposures, piericidin A exposures caused significant reductions in tPMP-1-induced killing of clinical strain 6850. Exposure of the tPMP-1r strains to piericidin A did not impact their baseline susceptibility profiles to tPMP-1 (Fig. 11A). Of note, both piericidin A as well as CCCP caused a significant reduction in tPMP-1-induced killing of clinical strain 6850 (Fig. 11B).
FIG. 10.
Impact of piericidin A treatment of tPMP-1s parental strains upon baseline tPMP-1 susceptibility profiles. Data represent the mean (±SD) of surviving CFU after tPMP-1 exposures (1 μg/ml) in the presence or absence of piericidin A (0, 4, or 8 μM). (A) Data for parental strain ISP479; (B) data for parental strain SH1000. Numbers above bars represent the mean fold increase in surviving CFU comparing piericidin A exposures to no-exposure controls. These data represent the means of at least two independent assays. *, not significantly different from untreated controls; **, P < 0.05 versus untreated controls.
FIG. 11.
(A) Demonstration of the impact of piericidin A treatment upon baseline tPMP-1 susceptibility profiles in two tPMP-1r strains. Numbers above bars represent the mean fold increases in surviving CFU comparing piericidin exposures to no-exposure controls. Data represent the mean (±SD) surviving CFU after tPMP-1 exposures (1 μg/ml) in the presence or absence of inhibitor. There were no significant changes in the presence of piericidin A. (B) Demonstration of the impact of two PMF inhibitors upon the tPMP-1 susceptibility profiles for clinical strain 6850. Numbers above bars represent the mean fold increases in surviving CFU comparing inhibitor exposures to no-exposure controls. Data represent the mean (±SD) of surviving CFU after tPMP-1 exposures (1 μg/ml) in the presence or absence of piericidin A (8 μM) or CCCP (100 μM). These data represent the means of at least two independent assays. *, P < 0.001; **, P < 0.0001.
DISCUSSION
Four phenotypes have been previously described as correlating with reduced in vitro susceptibility to tPMPs in S. aureus: (i) small-colony variants (SCVs), either selected by passage in gentamicin (20) or induced by mutagenesis of the hemB or menD locus (genes encoding components of the cytochrome and menaquinone pathways, respectively) (20). SCVs maintain a substantially lowered transmembrane potential than parental strains because of defects in their electron transport chain, especially during late log-stationary growth phases (4). Importantly, for cationic antimicrobial molecules such as aminoglycosides, a threshold ΔΨ appears to be critical for interacting with and transport through target microbial membranes. This concept underlies the common finding of reduced aminoglycoside susceptibilities in vitro for SCVs (4, 20). Of interest, SCVs also demonstrate reduced susceptibility to tPMPs, although a reduced ΔΨ is not the sole determinant of the tPMP-1r phenotype in vitro (20). (ii) Another phenotype is variants selected following serial passages (>10) of tPMP-1r S. aureus strains in increasing concentrations of this peptide (49). Similar to SCVs, such passage-derived tPMP-1r isolates exhibited a substantially diminished ΔΨ (A. S. Bayer, M. R. Yeaman, H.-G. Sahl, D. Brar, and R. A. Proctor, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. A-106, p. 19, 1997). (iii) Strains with relative increases in cell surface positive charge are a third phenotype. Peschel et al. have recently observed that the CM and cell walls of S. aureus contain a high proportion of positively charged phospholipids and teichoic acids, respectively, rendering such strains relatively resistant to killing by a broad range of cationic antimicrobial peptides, including tPMP-1 (38, 39). For the CM and cell walls, lysinylation of phosphotidyl-glycerol (38) and d-alanylation of teichoic acid (39), respectively, appear to be responsible for this phenotype. The proposed mechanism by which such positively charged surface envelope species are associated with reduced susceptibility to cationic peptides is via a charge-based surface repulsion (39). Finally, (iv) strains carrying plasmid determinants for cation export are the fourth phenotype. Staphylococci carrying plasmids (e.g., pSK1) encoding PMF-dependent, cation export pumps of the major facilitator superfamily (e.g., QacA) also exhibit reduced susceptibility to tPMP-1 in vitro, compared to isogenic plasmid-cured or qacA mutant constructs (26). The mechanism by which qacA expression confers reduced susceptibility to tPMPs remains unclear, although we have recently shown that tPMP-1 export does not play a role. (L. I. Kupferwasser et al., Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2288, 2000).
Despite distinct derivation strategies, strains with reduced susceptibilities to tPMP-1 (including the mutants in the current study) share selected phenotypic characteristics. We have previously shown that tPMP-1r strains exhibit increased membrane fluidity compared to their respective parental strains (5). The current study extends this observation, in that all the tPMP-1r transposon mutants demonstrated increased membrane fluidity. Also, similar to SCVs and tPMP-1r strains created by serial passage, the current transposon mutant strains all exhibited a reduced ΔΨ compared to their respective parental strains. Thus, both the parental strain and tPMP-1r constructs were able to maintain their initial ΔΨ due to the rapid consumption of glucose and excretion of lactic acid in a minimal medium. Ostensibly, lactic acid is then taken up by these cells under microaerophilic conditions and then completely oxidized (4, 42). However, strains with impaired electron transport (e.g., SCVs and complex I mutants) cannot subsequently maintain their ΔΨ (4).
Substantiating these concepts, preliminary data from tPMP-1r strains (compared to their respective parental strains) have two common CM features: (i) shorter fatty acid acyl chain length and (ii) increases in anteiso:iso branched chain fatty acid ratios (K. Mukhopadhyay et al., Abstr. 105th Gen. Meet. Am. Soc. Microbiol., abstr. A-002, p. 1, 2005). These latter parameters have been correlated with increased membrane fluidity in B. subtilis, particularly as part of the organism's response to cold shock (19). Of importance, increased membrane fluidity in biologic membranes has also been associated with excess proton leak, which itself contributes to a loss of ΔΨ (7). In addition, extremes of CM fluidity or rigidity are likely to impact the interactions of antimicrobial peptides with the target membrane (28). It should be emphasized that relatively small changes in CM characteristics (such as fluidity) have a major impact upon interactions with antimicrobial peptides (40).
The availability of a series of isogenic tPMP-1r strains provided a unique opportunity to further characterize the genetic basis for the tPMP-1r phenotype. Sequence analysis of chromosomal DNA flanking the ends of the transposon in tPMP-1r strains revealed that the transposon mutation resides within a gene encoding a member of the NADH-ubiquinone/plastoquinone (complex I) protein family. Complex I is a large, multisubunit respiratory chain enzyme that catalyzes the oxidation of NADH to NAD+, the reduction of quinone to quinol, and the transmembrane transfer of H+. Thus, this enzyme is critical in establishing the proton gradient and the PMF, especially under microaerophilic conditions and/or when linked to nitrate reductase. Three genes encoding complex I subunits in Escherichia coli, nuoL, nuoM, and nuoN, have homology with established Na+ (K+)/H+ antiporters. From an evolutionary perspective, it has been postulated that these subunits were ancestral antiport modules recruited to complex I (30, 31). Complex I-like antiporters have been identified in genome databases from diverse organisms and have been shown to function in the transmembrane exchange of Na+, K+, cholate, and H+ (3, 14, 17, 22, 23, 30, 41).
Staphylococcal snoD, like mrpD of Bacillus subtilis, resides in a seven-gene operon (17). These seven genes encode homologous proteins, and the overall architecture of the two operons is identical (17). In addition to a putative Na+ and cholate efflux pump encoded by mrpC and a solute-dependent pump encoded by mrpF, the products of the mrp operon appear to be required components of a redox-energized transporter in B. subtilis (18). The genes immediately upstream and downstream of the sno operon in S. aureus (SA0814 and SA0806, respectively) differ from those flanking mrp in B. subtilis. SA0814 is predicted to encode kinase-associated protein B, a B. subtilis kinB homologue, while SA0806 encodes a pyridine nucleotide-disulfide oxidoreductase family member (pfam 00070) with homology to a flavoprotein that is involved in K+ transport (GenBank accession no. 53736091). In B. subtilis, mpr is flanked by purT and ybfJ (a gene of unknown function that lacks homology with SA0806). The divergence in the sequences flanking the two loci suggest that disruption of sno operon function (rather than that of adjacent genes) is sufficient to confer the observed phenotypes associated with the tPMP-1r strains.
To verify the phenotypic consequences of the sno mutation, a wild-type copy of sno was reintroduced into the chromosome of several mutant constructs. This resulted in full restoration of parental-level protamine susceptibility to mutant strains, but only partial restoration of the tPMP-1s phenotype. The lack of full reconstitution of tPMP-1 susceptibility upon complementation is not unexpected and has been seen in other investigations of genetic loci involved in cationic peptide resistance (38, 39). Such lack of full phenotypic complementation has been variably ascribed to (i) plasmid copy number, (ii) plasmid stability, (iii) chromosomal integration of wild-type genes into a “nonnative” genetic context (i.e., into the lipase locus), or (iv) gene dosage effects from two or more copies of a gene of interest in the restored operon. While the latter two explanations apply to the current study, complementation of the sno operon may also be affected by the interference of products from a second sno-like gene identified in the genome of S. aureus N315 (SA0579 through SA0585). SnoD may encode the only gene product affected by the transposon mutation. However, we recognize that the expression of genes both upstream and downstream of snoD may have altered expression secondary to transposon disruption. Presently, we are examining the transcriptional capacity of the sno operon and creating a series of nonpolar deletion mutations in each of its seven genes to adjudicate these possibilities.
As noted before, the sno (mnh) operon was previously ascribed Na+/H+ antiporter function (15). However, our current data are more consistent with sno encoding a redox-energized complex involved in PMF maintenance, rather than a secondary Na+/H+ antiporter. In their original description of the mnh (sno) operon, Hiramatsu et al. (15) showed that an E. coli strain devoid of its major native Na+/H+ antiporters and which harbored a plasmid containing the sno operon was better able to survive an NaCl challenge. However, these data are also compatible with sno encoding a primary complex I-like enzyme system that supports Na+ transport, a possibility not addressed in the previous publication. In addition, these same investigators found that sno-encoded Na+ transport was susceptible to the PMF inhibitor CCCP. While prototypical secondary Na+/H+ antiporters are sensitive to H+ concentrations and therefore are PMF dependent, these data also support sno encoding a primary antiporter (e.g., energy providing, reaction coupled) (3, 24). Moreover, in our investigations, tPMP-1r constructs only exhibited a reduction in growth at high levels of NaCl, a finding also consistent with sno encoding a primary, not a secondary, Na+/H+ antiporter. Similarly, there were no differences in the growth kinetics of the mutant strains compared to the parental strains when grown under alkaline conditions. Secondary Na+/H+ antiporters would be expected to maximally function at alkaline pH. Finally, the tPMP-1r phenotype could be mimicked in both tPMP-1s parental strains by blocking the PMF with CCCP as well as by using the complex I inhibitor piericidin A.
Collectively, the above data strongly suggest that the reduced susceptibility to tPMP-1 and protamine in our transposon and transductant mutants involves disruption of a complex I-like enzyme locus. Moreover, these findings further strengthen our prior data, which emphasize the pivotal relationship between maintenance of a threshold ΔΨ and optimal in vitro killing of S. aureus by tPMP-1 (20, 21, 48).
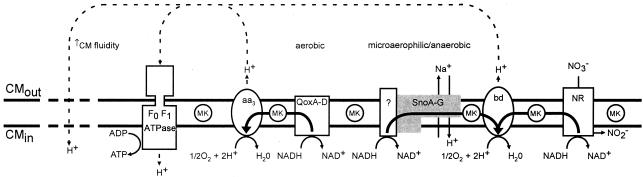
The electron transport systems of S. aureus as of yet are poorly defined. However, data available from the complete genomic sequences of this organism and from the current study suggest there are at least two distinct enzyme complexes that oxidize NADH and form part of the electron transport chain, QoxA-D and SnoA-G. Qox is a quinone oxidase that functions most efficiently under aerobic conditions (53). This gene is intact in snoD mutants; thus, these strains can transfer electrons to molecular oxygen using a mechanism that depends upon Qox complexes. However, the Sno complex becomes the predominant NADH oxidoreductase under microaerophilic conditions, when substrates and/or oxygen become limiting and when nitrate is used as the terminal electron acceptor. Both the enzymes above can generate a PMF via creation of the proton gradient, with the activity and efficiency of each system being dependent on substrate and oxygen availabilities. Our ΔΨ data support a key role for snoA-G in the generation of a PMF; however, further studies will be needed to delineate the net role of the Sno complex in the oxidation of NADH and electron transport. In Fig. 12 we present a hypothetical model of the role of SnoA-G within the electron transport chain of S. aureus and comment upon the impact of SnoA-G on tPMP susceptibilities.
FIG. 12.
Hypothetical model of the role of SnoA-G in electron transport in S. aureus. Depicted are the CM-associated components of a hypothetical electron transport chain: menaquinone (MK), the ATP synthase (F0 F1 ATPase), an aa3-type cytochrome (aa3), quinol oxidase (QoxA-D), the staphylococcal nuo-like orthologue complex (SnoA-G) coupled to an unidentified protein or proteins with NADH oxidase activity, a bd-type cytochrome, and nitrate reductase (NR). Reactions catalyzed by constituents of the hypothetical electron transport chain are shown in association with proteins or protein complexes. The thick arrows illustrate the flow of hydrogen ions (H+; dashed arrow). The aa3-type cytochromes are used in aerobic metabolism and are most closely linked to QoxA-D, an aerobic NADH oxidase. The bd-type cytochromes are used in microaerophilic and anaerobic metabolism and are linked to SnoA-G. Reduced susceptibility of S. aureus to tPMP-1 is predicted to be due to (i) the reduction in ΔΨ due to the loss of H+ translocation from the inside to the outside of the bacterium, from decreased electron flow to cytochrome bd, and (ii) an increase in diffusion of H+ from the outside to the inside of the bacterium caused by disordering (increased fluidity; dashed line) of the cytoplasmic membrane due to a shift from longer to shorter fatty acids and an increase in branched fatty acids.
Acknowledgments
We thank Jose Candelaria for excellent technical assistance in screening the transductant library and in the membrane fluidity assays. We also thank Nicole Baumert for performing the membrane potential assay. We thank Rajendra Prasad (J Nehru University, New Delhi, India) for assistance with the membrane fluidity assays and many helpful suggestions.
This research was supported in part by grants from the National Institutes of Health (AI053235-01 to P.J.M., AI42072 to R.A.P., AI-39108 to A.S.B., AI-47441 to A.L.C., and AI-48031 to M.R.Y.) H.-G.S. received financial support through the BONFOR program for Medical Faculty at the University of Bonn, Bonn, Germany.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. A., R. Brent, R. E. Kingston, R. E. Moore, D. D. Steidman, J. G. Smith, J. A. Smith, and K. Struhl (ed.). 1996. Current protocols in molecular biology. Greene Publishing and Wiley Interscience, New York, N.Y.
- 3.Barquera, B., P. Hellwig, W. Zhou, J. E. Morgan, C. C. Hase, K. K. Gosink, M. Nilges, P. J. Bruesehoff, A. Roth, C. R. Lancaster, and R. B. Gennis. 2002. Purification and characterization of the recombinant Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 41:3781-3789. [DOI] [PubMed] [Google Scholar]
- 4.Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8:253-260. [DOI] [PubMed] [Google Scholar]
- 5.Bayer, A. S., R. P. Prasad, J. Chandra, A. Koul, M. Smriti, A. Varma, R. A. Skurray, N. Firth, M. H. Brown, S.-P. Koo, and M. R. Yeaman. 2000. In vitro resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein in associated with alterations in cytoplasmic membrane fluidity. Infect. Immun. 68:3548-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertsova, Y., and A. V. Bogachev. 2004. The origin of the sodium-dependent NADH oxidation by the respiratory chain of Klebsiella pneumoniae. FEBS Lett. 563:207-212. [DOI] [PubMed] [Google Scholar]
- 7.Caiveau, O., D. Forune, S.-P. Cantrel, A. Zachowski, and F. Moreau. 2001. Consequences of omega-6-oleate desaturase deficiency on lipid dynamics and functional properties of mitochondrial membranes of Arabidopsis thaliana. J. Biol. Chem. 276:5788-5794. [DOI] [PubMed] [Google Scholar]
- 8.Coen, D. 1992. Enzymatic amplification of DNA by PCR: standard procedures and optimization, p. 15.1.1-15.1.7. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing and Wiley Interscience, New York, N.Y. [DOI] [PubMed]
- 9.Dhawan, V. K., A. S. Bayer, and M. R. Yeaman. 1998. Influence of in vitro susceptibility to thrombin-induced platelet microbicidal protein on the progression of experimental Staphylococcus aureus endocarditis. Infect. Immun. 66:3476-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhawan, V., M. R. Yeaman, E. Kim, A. L. Cheung, P. M. Sullam, and A. S. Bayer. 1997. Phenotypic resistance to thrombin-induced platelet microbicidal protein in vitro correlates with enhanced virulence in experimental endocarditis due to Staphylococcus aureus. Infect. Immun. 65:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyer, D. W., and J. J. Iandolo. 1983. Rapid isolation of DNA from Staphylococcus aureus. Appl. Environ. Microbiol. 46:283-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fowler, V. G., L. M. McIntyre, M. R. Yeaman, G. E. Peterson, L. B. Reller, G. R. Corey, and A. S. Bayer. 2000. In vitro resistance to thrombin-induced platelet microbicidal protein in isolates of Staphylococcus aureus from endocarditis patients correlates with an intravascular device. J. Infect. Dis. 182:1251-1254. [DOI] [PubMed] [Google Scholar]
- 13.Fowler, V. G., G. Sakoulas, L. M. McIntyre, V. Meka, R. Arbeit, C. H. Cabell, M. E. Stryjewski, G. M. Eliopoulos, L. B. Reller, G. R. Corey, T. Jones, N. Lucindo, M. R. Yeaman, and A. S. Bayer. 2004. Persistent bacteremia in methicillin-resistant Staphylococcus aureus is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190:1140-1149. [DOI] [PubMed] [Google Scholar]
- 14.Hamamoto, T., M. Hashimoto, M. Hino, M. Kitada, Y. Seto, T. Kudo, and K. Horikoshi. 1994. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol. Microbiol. 14:939-946. [DOI] [PubMed] [Google Scholar]
- 15.Hiramatsu, T., K. Kodama, T. Kuroda, T. Mizushima, and T. Tsuchiya. 1998. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J. Bacteriol. 180:6642-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, M., A. A. Guffanti, B. Oudegam, and T. A. Krulwich. 1999. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 181:2394-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, M., A. A. Guffanti, and T. A. Krulwich. 2001. Mrp-dependent Na+/H+ antiporters of Bacillus exhibit characteristics that are unanticipated for completely secondary active transporters. FEBS Lett. 496:117-120. [DOI] [PubMed] [Google Scholar]
- 19.Klein, W., M. H. Weber, and M. A. Marahiel. 1999. Cold shock response of Bacillus subtilis: isoleucine-dependent switch in the fatty acid branching pattern for membrane adaptation to low temperatures. J. Bacteriol. 181:5341-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koo, S.-P., A. S. Bayer, R. A. Proctor, H.-G. Sahl, and M. R. Yeaman. 1996. Staphylocidal action of platelet microbicidal protein is not solely dependent on intact transmembrane potential. Infect. Immun. 60:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koo, S.-P., B. L. Kagan, A. S. Bayer, and M. R. Yeaman. 1999. Membrane permeabilization by thrombin-induced platelet microbicidal protein-1 is modulated by transmembrane voltage orientation and magnitude. Infect. Immun. 67:2475-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosono, S., S. Morotomi, M. Kitada, and T. Kudo. 1999. Analyses of a Bacillus subtilis homologue of the Na+/H+ antiporter gene which is important for pH homeostasis of alkaliphilic Bacillus sp. C-125. Biochim. Biophys. Acta 1409:171-175. [DOI] [PubMed] [Google Scholar]
- 23.Kosono, S., Y. Ohashi, F. Kawamura, M. Kitada, and T. Kudo. 2000. Function of a principal Na+/H+ antiporter, ShaA, is required for initiation of sporulation in Bacillus subtilis. J. Bacteriol. 182:898-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krulwich, T. A., M. Ito, and A. A. Guffanti. 2001. The Na+-dependence of alkaliphily in Bacillus. Biochim. Biophys. Acta 1505:158-168. [DOI] [PubMed] [Google Scholar]
- 25.Kupferwasser, L. I., M. R. Yeaman, S. M. Shapiro, C. C. Nast, and A. S. Bayer. 2002. In vitro susceptibility to thrombin-induced platelet microbicidal protein is associated with reduced disease progression and complication rates in experimental Staphylococcus aureus endocarditis: microbiological, histopathological and echocardiographic anlyses. Circulation 105:746-752. [DOI] [PubMed] [Google Scholar]
- 26.Kupferwasser, L. I., R. A. Skurray, M. H. Brown, N. Firth, M. R. Yeaman, and A. S. Bayer. 1999. Plasmid-mediated resistance to thrombin-induced platelet microbicidal protein in staphylococci: role of the qacA locus. Antimicrob. Agents Chemother. 43:2395-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 28.Lohner, K., and S. E. Blondelle. 2005. Molecular mechanisms of membrane perturbation by antimicrobial peptides and the use of biophysical studies in the design of novel peptide antibiotics. Combin. Chem. High Throughput Screening 8:239-255. [DOI] [PubMed] [Google Scholar]
- 29.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathiesen, C., and C. Hagerhall. 2003. The “antiporter module” of respiratory chain complex I includes the MrpC/NuoK subunit—a revision of the modular evolution scheme. FEBS Lett. 549:7-13. [DOI] [PubMed] [Google Scholar]
- 31.Mathiesen, C., and C. Hagerhall. 2002. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta 1556:121-132. [DOI] [PubMed] [Google Scholar]
- 32.McNamara, P. J., and J. J. Iandolo. 1998. Genetic instability of the global regulator agr explains the phenotype of the xpr mutation in Staphylococcus aureus KSI9051. J. Bacteriol. 180:2609-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell, B. A., I. T. Paulsen, M. H. Brown, and R. A. Skurray. 1999. Bioenergetics of the staphylococcal multidrug export protein QacA: identification of distinct binding sites for monovalent and divalent cations. J. Biol. Chem. 274:3541-3548. [DOI] [PubMed] [Google Scholar]
- 34.Nesin, M., P. Svec, J. R. Lupski, G. N. Godson, B. Kreiswirth, J. Kornblum, and S. J. Projan. 1990. Cloning and nucleotide sequence of a chromosomally encoded tetracycline resistance determinant, tetA(M), from a pathogenic, methicillin-resistant strain of Staphylococcus aureus. Antimicrob. Agents Chemother. 34:2273-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 36.Palma, M., and A. L. Cheung. 2001. sigmaB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pattee, P. A. 1981. Distribution of Tn551 insertion sites responsible for auxotrophy on the Staphylococcus aureus chromosome. J. Bacteriol. 145:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel, and J. A. van Strijp. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 40.Pokorny, A., and P. F. F. Almeida. 2004. Kinetics of dye efflux and lipid flip-flop induced by delta-lysin in phosphatidyl vesicles and the mechanism of graded release by amphipathic, α-helical peptides. Biochemistry 43:8846-8857. [DOI] [PubMed] [Google Scholar]
- 41.Putnoky, P., A. Kereszt, T. Nakamura, G. Endre, E. Grosskopf, P. Kiss, and A. Kondorosi. 1998. The pha gene cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol. Microbiol. 28:1091-1101. [DOI] [PubMed] [Google Scholar]
- 42.Sahl, H. G. 1985. Influence of the staphylococcin-like peptide Pep 5 on membrane potential of bacterial cells and cytoplasmic membrane vesicles. J. Bacteriol. 162:833-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang, Y. Q., M. R. Yeaman, and M. E. Selsted. 2002. Antimicrobial peptides from human platelets. Infect. Immun. 70:6524-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, T., M. R. Yeaman, and A. S. Bayer. 1994. Resistance to platelet microbicidal protein in vitro among bacteremic staphylococcal and viridans streptococcal isolates correlates with an endocarditis source. Antimicrob. Agents Chemother. 38:729-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong, Y. Q., A. S. Bayer, and M. R. Yeaman. 2002. Inhibition of intracellular macromolecular synthesis in Staphylococcus aureus by thrombin-induced platelet microbicidal proteins. J. Infect. Dis. 185:348-356. [DOI] [PubMed] [Google Scholar]
- 46.Xiong, Y. Q., M. R. Yeaman, and A. S. Bayer. 1999. In vitro antibacterial activities of platelet microbicidal protein and neutrophil defensin against Staphylococcus aureus are influenced by antibiotics differing in mechanism of action. Antimicrob. Agents Chemother. 43:1111-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeaman, M. R. 1997. The role of platelets in antimicrobial host defense. Clin. Infect. Dis. 25:951-970. [DOI] [PubMed] [Google Scholar]
- 48.Yeaman, M. R., A. S. Bayer, S.-P. Koo, W. Foss, and P. M. Sullam. 1998. Platelet microbicidal protein and neutrophil defensin disrupt the Staphylococcus aureus cytoplasmic membrane by distinct mechanism of action. J. Clin. Investig. 101:178-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeaman, M. R., D. C. Norman, and A. S. Bayer. 1992. Platelet microbicidal protein enhances antibiotic-induced killing of and postantibiotic effect in Staphylococcus aureus. Antimicrob. Agents Chemother. 36:1665-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeaman, M. R., Y.-Q. Tang, A. J. Shen, A. S. Bayer, and M. E. Selsted. 1997. Purification and in vitro activities of rabbit platelet microbicidal proteins. Infect. Immun. 65:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yount, N. Y., K. D. Gank, Y. Q. Xiong, A. S. Bayer, T. Pender, W. H. Welch, and M. R. Yeaman. 2004. Platelet microbicidal protein-1: structural themes of a multifunctional antimicrobial peptide. Antimicrob. Agents Chemother. 48:4395-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yount, N. Y., and M. R. Yeaman. 2004. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA 101:7363-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zamboni, N., and U. Sauer. 2003. Knockout of the high-coupling cytochrome aa3 oxidase reduces TCA cycle fluxes in Bacillus subtilis FEMS Microbiol. Lett. 226:121-126. [DOI] [PubMed] [Google Scholar]