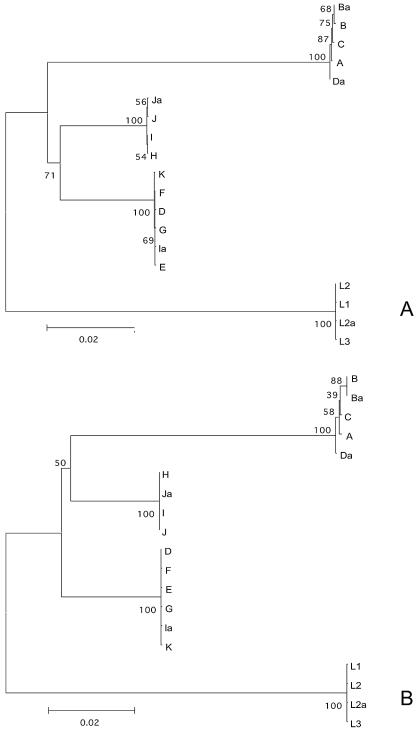
FIG. 2.
Phylogenetic reconstruction of the nucleotide (A) and amino acid (B) sequences showing the evolutionary history of pmpF by neighbor-joining tree topologies, based on distance estimates using a Kimura two-parameter model for substitution events (nucleotides) and the gamma distance model (amino acids). These reconstructions are based on pmpF sequences of the 18 serovars and the genovariant Ja of C. trachomatis. Branch lengths are proportional to distances between serovars. The values at the nodes are the bootstrap confidence levels representing the percentage of 1,000 bootstrap replicates for which the serovar to the right was separated from the others.