Abstract
Topoisomerase I is a ubiquitous DNA-cleaving enzyme and an important therapeutic target in cancer chemotherapy for camptothecins (CPTs). These drugs stimulate DNA cleavage by topoisomerase I but exhibit little sequence preference, inducing toxicity and side effects. A convenient strategy to confer sequence specificity consists of the linkage of topoisomerase poisons to DNA sequence recognition elements. In this context, triple-helix-forming oligonucleotides (TFOs) covalently linked to CPTs were investigated for the capacity to direct topoisomerase I-mediated DNA cleavage in cells. In the first part of our study, we showed that these optimized conjugates were able to regulate gene expression in cells upon the use of a Photinus pyralis luciferase reporter gene system. Furthermore, the formation of covalent topoisomerase I/DNA complexes by the TFO-CPT conjugates was detected in cell nuclei. In the second part, we elucidated the molecular specificity of topoisomerase I cleavage by the conjugates by using modified DNA targets and in vitro cleavage assays. Mutations either in the triplex site or in the DNA duplex receptor are not tolerated; such DNA modifications completely abolished conjugate-induced cleavage all along the DNA. These results indicate that these conjugates may be further developed to improve chemotherapeutic cancer treatments by targeting topoisomerase I-induced DNA cleavage to appropriately chosen genes.
Topoisomerase I (Topo I) is a ubiquitous nuclear enzyme involved in the control of DNA topology. During the catalytic cycle, the enzyme transiently cleaves DNA and forms a covalent 3′-phosphorotyrosyl adduct usually referred to as the cleavage complex (14, 28, 29). A number of drugs, such as the antitumor alkaloid camptothecin (CPT) (for reviews, see references 17 and 24) and indolocarbazole analogs of the antibiotic rebeccamycin family (9, 23, 30), act by blocking the religation step after DNA cleavage, thereby enhancing the formation of cleavage complexes (DNA/Topo I/drug ternary complexes), which constitute persistent DNA breaks believed to be responsible for cell death (20-22, 28). Topo I poisons stimulate DNA cleavage at many sites along the double helix with a limited sequence preference (one or two bases around the cleavage site). However, the sequence specificity of the Topo I poison can be considerably enhanced by linkage to a DNA recognition element. Initially, Matteucci et al. (19) showed in vitro that the covalent attachment of CPT to a triple-helix-forming oligonucleotide (TFO) induces site-specific DNA cleavage near the end of the triple helix. In the same context, we showed that CPT and rebeccamycin derivatives covalently linked to a TFO induce Topo I to mediate a sequence-specific DNA cleavage near the triplex site (3, 6, 7) according to a preferred geometry (2). The use of triple-helical DNA structures offers an efficient strategy to target Topo I to specific DNA sequences. Consequently, TFO-drug conjugates may be potentially exploited to improve the efficacy and selectivity of chemotherapeutic cancer treatments with Topo I poisons by targeting specific genes and thus reducing drug toxicity.
Here we evaluated the ability of the TFO-CPT conjugates to recruit Topo I in cells. By using a luciferase expression system containing the triplex sequence in the 5′ transcribed region, we showed that the TFO-CPT conjugates inhibited gene expression more efficiently than the triplex alone. Finally, DNA/Topo I complexes were detected in cell nuclei after treatment with TFO-CPT conjugates. The mechanism of targeting was further elucidated by using modified targets for in vitro cleavage assays; our results showed that in order to realize efficient cleavage, the TFO moiety must bind to the duplex and a topoisomerase cleavage site must be present in close proximity to the triplex site where the CPT is located. The same requirements were necessary for cellular activity.
MATERIALS AND METHODS
Spectral analysis.
For the cCPT (10-carboxymethyloxy-CPT) derivative, mass determination was accomplished by electrospray ionization on a Finnigan 3200 Quadrupole mass spectrometer. High-performance liquid chromatography (HPLC) purifications were performed upon a Varian Associates HPLC apparatus with an Altech Alltima C18 reversed-phase column (250 by 10 mm, 5 μm). 1H and 13C nuclear magnetic resonance spectra were recorded in chloroform-d, acetone-d6, or dimethyl sulfoxide (DMSO)-d6 on a Varian spectrometer (300 MHz). For all oligonucleotide conjugates, mass determination was accomplished by electrospray ionization on a Q-STAR pulsar I (Appleura) and HPLC purifications were performed upon an Agilent 1100 with an Xterra reversed-phase C18 column (4.6 by 50 mm, 2.5 μm). Absorbance spectrophotometry was performed on a Uvikon 860 (Kontron).
Materials.
All chemicals were purchased from Aldrich Chemical Company. All solvents were of analytical grade. All synthetic transformations of CPT were carried out under dry argon or nitrogen.
MGB hexa(N-methylpyrrole)carboxamide was obtained as previously described (5).
CPTs.
10-Hydroxy-CPT was purchased from Sino Dragon I/E Co. Ltd., 10CPT (10-carboxy-CPT) and 7CPT [7-(2-aminoethyl)-CPT] were obtained as previously described (7), and cCPT was synthesized by a slight modification of the procedure described in reference 27. CPT analogs were dissolved in DMSO at 3 mg/ml and then diluted further with water. The final DMSO concentration never exceeded 0.3% (vol/vol) in the assays. Each CPT analog was attached to the 3′ end of the TFO as shown in Fig. 1B.
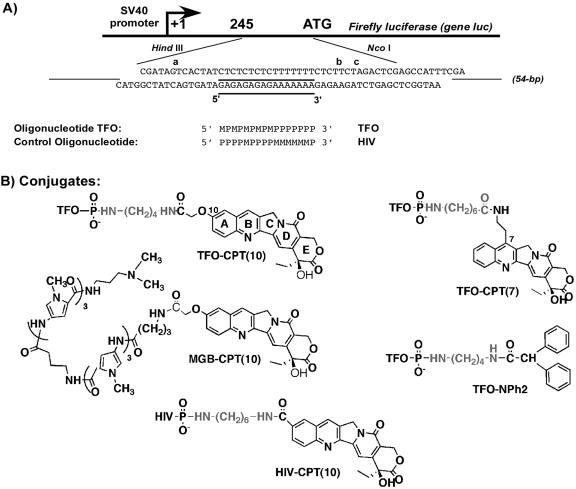
FIG. 1.
(A) Sequences of the TFOs and of the duplex containing the 16-bp (underlined) triplex target site used in this study. The experimental construction for cell experiments is shown: an insert of 54 bp, containing the original triplex target (plasmid pWT), was cloned within the 5′ transcribed but untranslated region of the firefly luciferase gene (luc), under the control of the SV40 promoter, between the HindIII and NcoI sites of vector pGL3Pr. The 54-bp region around the triplex site is shown. The region on the 3′ side of the triple helix, i.e., the 3′ side of the oligopurine strand of the duplex, contains two Topo I-mediated cleavage sites, b and c, observed in the presence of free CPT. All Topo I cleavage sites observed in vitro are indicated by letters. M, 5-methyl-2′-deoxycytidine; P, 5-propynyl-2′-deoxyuridine. The 16-nt sequence of the HIV-CPT(10) conjugate used as a control is shown. (B) Structures of the CPT conjugate derivatives used in this study.
Oligonucleotides.
Oligonucleotides were purchased from Eurogentec and purified with quick-spin Sephadex G-25 (Bio-Rad). Concentrations were determined spectrophotometrically at 25°C with molar extinction coefficients at 260 nm calculated from a nearest-neighbor model (11).
The nomenclature of the oligonucleotides and conjugates is the following: the abbreviation TFO is followed by the designation of the poison and its attachment point [CPT(10) for CPT attached through position 10 upon the use of the derivative 10CPT or cCPT and CPT(7) for attachment through position 7 with the derivative 7CPT]. The orientation of the triple helix is defined as the orientation of the purine-rich strand of the duplex, and the TFO binds in the major groove in an orientation parallel to the oligopurine strand. All conjugates bear the CPT derivative at the 3′ end of the TFO and thus at the 3′ end of the triple-helical structure.
Synthesis of conjugates.
The CPT derivatives were conjugated to the terminal amino group of different linker arms at the 3′ end of the oligonucleotide as previously described (5, 7) or as follows. TFO-NH2 corresponds to the TFO attached to a linker before coupling to CPT analogs and purified. Human immunodeficiency virus (HIV)-CPT(10) and TFO-CPT(7) were synthesized and analyzed as described in reference 7.
Synthesis of TFO-NPh2.
Six hundred twenty micrograms (120 nmol) of 3′-phosphorylated oligonucleotide was precipitated as the hexadecyltrimethylammonium salt and then dissolved in 50 μl of dry N,N-dimethylformamide. Solutions of 4-N,N-dimethylaminopyridine (5 mg in 25 μl of DMSO, 41 μmol), dipyridyl disulfide (6.6 mg in 25 μl of DMSO, 30 μmol), and triphenylphosphine (7.9 mg in 50 μl of DMSO, 30 μmol) were added. After 15 min of incubation at room temperature, 1,4-diaminobutane (5 mg, 57 μmol) was added to the activated oligonucleotide. The mixture was kept for 2 h at room temperature. The oligonucleotide derivative was then precipitated with 2% LiClO4 in acetone, rinsed with acetone, and dissolved in 50 μl of water. The attachment of the diamino group was quantitative. Then, diphenylacetic acid was attached by activating the carboxyl group with HATU. The oligonucleotide derivative was precipitated as the hexadecyltrimethylammonium salt and then dissolved in 100 μl of dry N,N -dimethylformamide. Diphenylacetic acid (1 mg, 5 mmol), HATU (1.9 mg, 5 mmol), and 1 μl of triethylamine were added. The mixture was kept for 4 h at room temperature. The oligonucleotide conjugate was then precipitated with 2% LiClO4 in acetone, rinsed with acetone, and purified by reversed-phase HPLC with a linear acetonitrile gradient (0→80% CH3CN in 0.2 M ammonium acetate). The average yield was 60%. The oligonucleotide conjugates were characterized by UV spectroscopy, denaturing gel electrophoresis, and mass spectrometry. {TFO-NPh2 MS (ES−) m/z: 5,406 [M-H]− (calculated: 5,408).}
Synthesis of TFO-CPT(10).
HATU was used for the coupling reaction with cCPT, replacing the previously used procedure (2). Three hundred ten micrograms (60 nmol) of 3′-phosphorylated TFO was precipitated as the hexadecyltrimethylammonium salt and dissolved in 50 μl of dry DMSO. 4-N,N-Dimethylaminopyridine (5 mg in 25 μl of DMSO, 41 μmol), dipyridyl disulfide (6.6 mg in 25 μl of DMSO, 30 μmol), and triphenylphosphine (7.9 mg in 50 μl of DMSO, 30 μmol) were added, and after 15 min of incubation at room temperature, 1,4-diaminobutane (5 mg, 57 μmol) was added. After a 2-h incubation at room temperature, the oligonucleotide derivative was precipitated with 2% LiClO4 in acetone and dissolved in 50 μl of water. The attachment of the diamino linker was quantitative. cCPT (400 μg, 1 μmol), HATU (380 μg, 1.1 μmol), and 1 μl of triethylamine were then added to the oligonucleotide derivative precipitated as the hexadecyltrimethylammonium salt and dissolved in 100 μl DMSO. The reaction mixture was kept at room temperature for 4 h. After precipitation with 2% LiClO4 in acetone, the oligonucleotide conjugate was purified by reversed-phase HPLC with a linear acetonitrile gradient (0→80% CH3CN in 0.2 M ammonium acetate). The average yield was 40%. The oligonucleotide conjugates were characterized by UV spectroscopy, denaturing gel electrophoresis, and mass spectrometry. {TFO-CPT(10) MS (ES−) m/z: 5,618 [M-H]− (calculated: 5,617).}
Synthesis of MGB conjugates.
cCPT was attached to the minor-groove binder MGB hexa(N-methylpyrrole)carboxamide as previously described (5). {MGB-CPT(10) MS (ES−) m/z: 1,409 [M-H]− (calculated: 1,409).}
Plasmids and DNA fragments.
Plasmid pBSK(+/−) was bought from Promega, and the four 77-bp target duplexes were inserted between its BamHI and EcoRI sites (Fig. 5 contains the sequences). Digestion of the plasmid by PvuII and EcoRI yielded a 324-mer fragment suitable for 3′-end labeling by the avian myeloblastosis virus reverse transcriptase (New England Biolabs) and [α-32P]ddATP(Amersham), used for Topo I cleavage assays. The procedures used for isolation, purification, and labeling of this duplex DNA fragment have been described previously (8).
FIG. 5.
Modified duplexes are derived from duplex WT by mutations (in bold) at the triplex site (MUT) or at cleavage sites b and c, replaced by either no cleavage site (B2) or by a known Topo I cleavage site, B (TID).
The plasmids used in luciferase expression assays were obtained from the pGL3Promoter (pGL3Pr) vector from Promega by cloning a 54-bp insert into the 5′ transcribed but untranslated region of the firefly (Photinus pyralis) luciferase gene (HindIII-NcoI region) under the control of the simian virus 40 (SV40) promoter. The two 54-bp duplexes contained either the intact wild-type (WT) target with the triplex (underlined) and CPT site (5′AGCTTTACCGAGCTCAGATCTTCTCTTTTTTTCTCTCTCTCTATCACTGATAGC3′/5′CATGGC TATCAGTGATAGAGAGAGAGAAAAAAAGAGAAGATCTGAGCTCGGTAA3′, plasmid pWT) or the triplex site without cleavages sites b and c (in bold) at its 3′ end (B2) (5′AGCTTTACCGAGCTCAGAAAAACTCTTTTTTTCTCTCTCTCTATCACTGATAGC3′/5′CATGGCTATCAGTGATAGAGAGAGAGA AAAAAAGAGTTTTTCTGAGCTCGGTAA3′, plasmid called pB2). The Renilla reniformis luciferase control vector pTK-RL from Promega was used to monitor transfection efficiencies. In this vector, Renilla luciferase expression was under the control of the herpes simplex virus thymidine kinase promoter region.
Topo I cleavage assays.
The radiolabeled target duplexes (50 nM) were incubated for 1 h at 30°C in 50 mM Tris-HCl (pH 7.5)-60 mM KCl-10 mM MgCl2-0.5 mM dithiothreitol -0.1 mM EDTA-30 μg/ml bovine serum albumin in the presence of the ligand and/or the drugs at the indicated concentration (total reaction volume, 10 μl). To analyze the Topo I DNA cleavage products, 10 U of calf thymus enzyme (Invitrogen) was added to the duplex and the mixture was preincubated as described above and then incubated for 15 min at 30°C. The DNA-Topo I cleavage complexes were dissociated by addition of sodium dodecyl sulfate (final concentration, 0.5%). After ethanol precipitation, all samples were resuspended in 6 μl of formamide, heated at 90°C for 4 min, and then chilled on ice for 4 min before being loaded onto a denaturing 8% polyacrylamide gel (19:1 acrylamide-bisacrylamide) containing 7.5 M urea in TBE buffer (50 mM Tris base, 55 mM boric acid, 1 mM EDTA). To quantitate the extent of cleavage, the gels were scanned with a Typhoon 9410 (Amersham Biosciences). For determination of cleavage levels, normalization relative to total loading was performed. The experiments were repeated between 4 and 10 times.
Triplex formation assay.
DNA triplex formation was followed by gel retardation as previously described (3). Increasing concentrations (100 nM to 10 μM) of the TFOs were added to a 10 nM concentration of the radiolabeled duplex in 10 mM MgCl2-100 mM NaCl-50 mM HEPES (pH 7.2)-10% sucrose-0.5 mg/ml tRNA with sample incubation at 37°C for 24 h. Electrophoresis was performed on a nondenaturing 12% polyacrylamide gel containing 10 mM MgCl2 and 50 mM HEPES, pH 7.2, at 37°C. The concentration at which 50% of the triplex was formed is reported as micromolar.
Cell cultures.
Human HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, and 1% penicillin-streptomycin, at 37°C in 5% CO2.
Transient-expression experiments. (i) Transfections.
Transfections were performed by using Superfect (QIAGEN). To each well of a 96-well plate containing 125 μl of cells at a concentration of 110,000/ml were added 112.5 μl of fresh serum-containing medium and 12.5 μl of transfection mixture, prepared according to the manufacturer's instructions with 1 μl of transfecting agent, 125 ng of plasmid pWT or control pGL3Pr, 62.5 ng of pTK-RL, and different amounts of TFO conjugates. The TFO conjugates were present in the wells during transfection at final concentrations of 0.05, 0.1, and 0.5 μM.
(ii) Luciferase assay.
Both luciferase activities (P. pyralis and R. reniformis) were measured in cell lysates by using the dual-luciferase reporter assay system from Promega. A luminometer (Victor; Wallac) was used to measure light signal output. The experiments were run as triplicates. The results are reported as the mean P. pyralis/R. reniformis luciferase ratio and are further normalized to the ratio of the cells not treated with the conjugates. The total protein amount was measured with the Bio-Rad protein assay according to the manufacturer's instructions. Experiments were repeated 3 to 10 times.
ICE (in vivo complex of enzyme) bioassay.
HeLa cell nuclei were prepared as described previously (15). The in vivo Topo I link kit of TopoGEN, Inc. (Columbus, OH), was used to determine the ability of the conjugates to induce Topo I-mediated DNA breaks in treated HeLa cell nuclei. The recommended protocol was followed, with a few modifications. HeLa cell nuclei (5 × 106) were prepared and incubated for 3 h at 37°C with free CPT, with CPT-oligonucleotide conjugates, or with a control oligonucleotide (TFO-NPh2) at various concentrations. Cells nuclei were pelleted by centrifugation (1,000 rpm, 5 min) and rapidly resuspended in 0.8 ml of the lysis buffer (10 mM Tris HCl, pH 7.5, 1 mM EDTA, 1% Sarkosyl). The lysed cell mixture was then overlaid onto a CsCl density gradient containing four different density steps (0.8 ml of CsCl at 1.82, 1.72, 1.50, and 1.37 g/ml). The tubes were centrifuged in a Beckman SW60 rotor at 31,000 rpm (13,000 × g) for 15 h at 25°C. From the top of the gradient, 12 fractions of 330 μl were collected. The DNA content in each fraction was estimated by absorbance measurement at 260 nm. For the immunoblot analysis, 50 μl of each fraction was diluted with 100 μl of 25 mM sodium phosphate buffer (PBS, pH 6.5) prior to applying the diluted solution to the slot blot unit under a mild vacuum. PBS-washed Hybond-C extra nitrocellulose membranes (Amersham) cut to fit the vacuum slot blot device Hybri-Slot 24 (Life Science, Cergy-Pontoise, France) were loaded with the diluted samples, washed briefly with PBS, and then soaked for 2 h in TBST (20 mM Tris HCl, pH 7.6, 137 mM NaCl, 0.1% Tween 20, 1% bovine serum albumin) supplemented with 5% nonfat dried milk. The membranes were washed three times (10 min per wash) with TBST prior to incubation for 1 h at room temperature with anti-Topo I antibody (1/10,000 dilution in 25 ml TBST; Santa Cruz Biotechnology Inc.). After three successive washes (10 min with TBST), the membranes were incubated with an anti-rabbit antibody conjugated to horseradish peroxidase (1/1,000 dilution in 25 ml TBST; Amersham) for 30 min. After four successive washes (10 min each with TBST), the ECL Western blotting detection reagents from Amersham were used for detection. Fractions 1 to 4 contain free Topo I. DNA-Topo I covalent complexes can be detected in fractions 8 to 11 of the drug-treated samples. To quantify the amount of cleavage complexes formed in the presence of free or conjugated CPT, the percentage of signal in fractions 8 to 11 was normalized to the total amount of Topo I signal in all fractions for each sample.
Stability of oligonucleotides in cell media.
The conjugate TFO-CPT(10) and precursor TFO-P were radiolabeled at the 5′ end with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Amersham). The radiolabeled oligonucleotides (0.05 μM) were incubated in cell culture medium (DMEM containing 10% fetal bovine serum) at 37°C. Aliquots were removed after 5 min (0 h), 24 h, or 72 h. Proteins were eliminated by a phenol-chloroform extraction procedure, and the aqueous phase was then ethanol precipitated. The degradation pattern (full-length and degraded products) was analyzed by gel electrophoresis on a 20% denaturing gel.
RESULTS
We investigated whether a TFO-CPT conjugate was able to induce the formation of a Topo I/DNA complex in a cellular context and its consequences. To do this, we constructed a P. pyralis luciferase reporter system containing a triplex target site, previously studied (7), and synthesized a series of TFO-CPTs.
Experimental design.
The WT target duplex contains a 16-bp oligopyrimidine-oligopurine sequence suitable for triplex formation (underlined in Fig. 1A). A 16-mer TFO, containing 5-methyl-2′-deoxycytidine (M) and 5-propynyl-2′-deoxyuridine (P), was used in order to form a stable pyrimidine motif triple helix at neutral pH (4, 16, 18). The cleavage sites induced by free CPT in the presence of Topo I are indicated by letters (a, b, and c). In this study, two TFO-CPT conjugates were used (Fig. 1B); cCPT was attached to the 3′ end of the TFO via a butyldiamino linker [TFO-CPT(10)], and 7CPT was attached through a carboxyhexylamine [TFO-CPT(7)]. The latter has been previously described, optimized for linker arm length, and characterized for its triplex-directed DNA cleavage by Topo I (7). The former differs from a previously synthesized active TFO-CPT conjugate by the absence of an ethyl chain in position 7 (2). The linker arm has been optimized for maximum cleavage in vitro. As a control, we chose to modify the TFO at its 3′ end by attaching a diphenyl-acetyl group (TFO-NPh2) via a butyldiamino linker [NH2-(CH2)4-NH2]. The diphenylacetyl group has been added in order to replace the primary amino group of the linker at the 3′ end of the TFO (TFO-NH2) with aromatic groups to mimic the polyaromatic CPT. This control TFO was used instead of directly using TFO-NH2, bearing only the linker arm, or the precursor TFO-P, the TFO phosphorylated at the 3′ end, because its triplex stability is comparable to that of TFO-CPT(10). In fact, the concentration at which 50% of the triplex is formed is 1.5 μM for TFO-CPT(10), 1.8 μM for TFO-NPh2, 0.6 μM for TFO-P, and 0.7 μM for TFO-NH2, as measured by gel shift assay. Another control oligonucleotide-CPT conjugate was synthesized in which the oligonucleotide part of the conjugate contains the same number of bases (16 nucleotides long) and the same M- and P-modified bases but a different sequence so that it cannot bind to the triplex target [called HIV-CPT(10); Fig. 1A and B show its sequence and chemical structure, respectively]. 10CPT and a hexyldiamino linker arm were used for this conjugate, which belongs to another project aimed at targeting the HIV type 1 genome. The hexyl linker arm was used in order to maintain nine atoms between position 10 on the CPT and the phosphate group at the 3′ end of the oligonucleotide, as in the case of conjugate TFO-CPT(10).
Cellular activity of TFO-CPT conjugates.
The experimental construction used is the following. A 54-bp duplex containing the triplex site and CPT cleavage sites a, b, and c was cloned at the HindIII/NcoI sites into the pGL3 Promoter vector (pGL3Pr) containing the P. pyralis luciferase gene under the control of the SV40 promoter (Fig. 1A). The TFO-binding sequence and a site sensitive to CPT in the vicinity thereof are thus placed in the transcribed region upward of the P. pyralis luciferase (luc) gene. The intact triple-helix sequence was inserted in such a way that the synthesized RNA contains the oligopyrimidine strand in order to avoid an antisense effect of the TFO. We verified that the TFO-CPT conjugate induced a sequence-specific Topo I-mediated DNA cleavage in vitro on this construct (see Fig. 1S in the supplemental material). As a control, compound HIV-CPT(10) was used to exclude an effect of the tethered CPT by itself, because the oligonucleotide part of the conjugate can no longer bind to the target duplex and thus cannot deliver the CPT on the duplex.
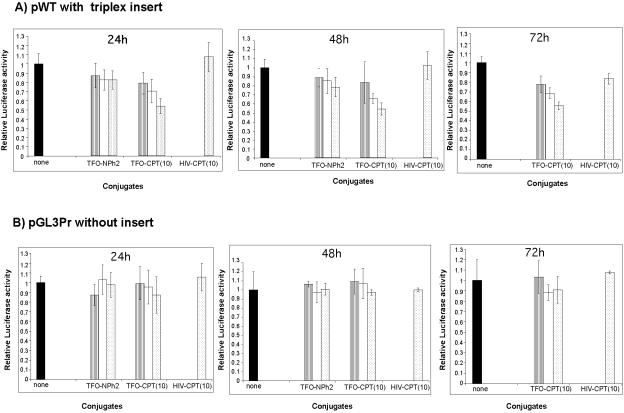
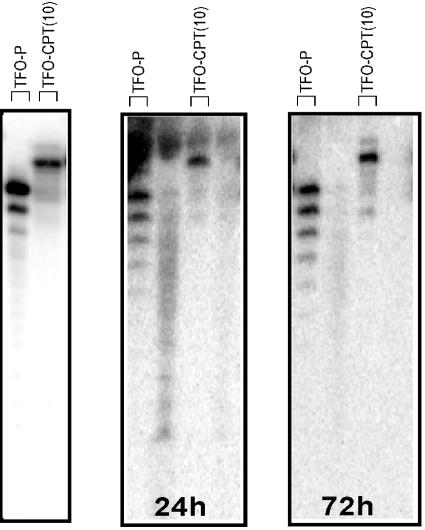
The expression plasmid vector obtained in this way was cotransfected with the expression vector for Renilla luciferase (pTK-RL, used as a transfection control) in the absence or presence of different conjugates at the indicated concentration. As the active conjugate, we chose TFO-CPT(10) and, as a reference, conjugate TFO-NPh2, in which the oligonucleotide is protected at the 3′ end by the linker arm and a diphenylacetyl group. The latter conjugate was chosen to evaluate the effect of the triple helix alone on the inhibition of transcription of the luc gene. At 24, 48, and 72 h after transfection with Superfect, the cells were lysed and tested for Pyralis and Renilla luciferase activities. The ratio of the luminescence intensities of the Pyralis and Renilla luciferases normalized to the untreated cells is represented in Fig. 2. Figure 2A illustrates, for the first time with TFO-CPT molecules, the inhibition of the expression of the target gene in HeLa cells. The conjugate TFO-CPT(10) reduced luciferase expression by about 40 to 50% at 0.5 μM; i.e., a residual luciferase activity of 60 to 50% was observed (Fig. 2A). An equivalent inhibition of 40% was observed with conjugate TFO-CPT(7) (data not shown). This inhibition effect lasted with time, and the same luciferase activity inhibition was measured at 48 and 72 h after transfection. In contrast, the reference (TFO-NPh2) and control [HIV-CPT(10)] oligonucleotides had no significant effect on the expression of firefly luciferase under our experimental conditions, up to 2 μM. Finally, with control plasmid pGL3Pr, which has no insert (and thus no triplex can be formed), the conjugate had no significant effect at 24, 48, or 72 h after transfection, nor did the control oligonucleotides (Fig. 2B). In our experiments, no increase in cell lethality (measured as the total protein level in the cell lysate) was observed upon transfection of the conjugates. It is noteworthy that the presence of the linker arm or of the CPT analog at the 3′ end of the oligonucleotides protects them from rapid degradation in cells, mainly by 3′ exonucleases, up to 72 h. In Fig. 3 is reported the analysis by gel sequencing of the 5′-radiolabeled TFO-P and TFO-CPT(10) (left panel) after incubation in cell culture medium for 24 h (central panel) or 72 h (right panel). While the TFO-P was partially degraded (62% of the original level of intact oligonucleotide was measured at 24 h and 54% at 72 h), 93% of the original level of intact TFO-CPT(10) was found at 24 h and 89% at 72 h.
FIG. 2.
Cellular activities of TFO-CPT conjugates. HeLa cell lysates were assayed for firefly and Renilla luciferase activities after transfection by Superfect. The firefly luciferase construct was introduced with the control pTK-RL vector in the absence (black bars) or presence of TFO-NPh2, TFO-CPT(10), or HIV-CPT(10) at 0.05 μM (hatched bars), 0.1 μM (white bars), or 0.5 μM (dotted bars). The results presented are the mean firefly/Renilla luciferase luminescence intensity ratios of triplicates (relative luciferase activity) and are normalized to the value obtained in the absence of the oligonucleotides (none). (A) pWT construct at 24 h, 48 h, and 72 h after transfection. (B) pGL3Pr control vector at 24 h, 48 h, and 72 h after transfection. Conjugates TFO-CPT(10) and TFO-CPT(7) gave equivalent results.
FIG. 3.
Stability of TFO-P and conjugate TFO-CPT(10) in cell medium. The 5′-end-radiolabeled oligonucleotides (left panel) were incubated in cell culture medium (DMEM containing 10% fetal bovine serum) at 37°C. Aliquots were removed after 24 h (central panel) or 72 h (right panel) and analyzed on a 20% denaturing gel.
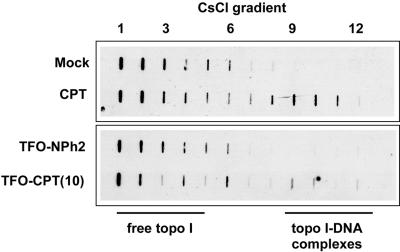
According to these results, the specific TFO-CPT conjugate directed against a triplex site situated upstream of the luc gene is able to inhibit luciferase expression more efficiently than the TFO-NPh2 conjugate lacking the CPT moiety. However, this experiment does not measure directly the ability of the TFO-CPT conjugates to induce the stabilization of the Topo I/DNA cleavage complex in cells. To address this issue, we used the ICE bioassay, which detects the formation of covalent Topo I/DNA complexes in cells. HeLa cell nuclei were incubated for 3 h at 37°C with CPT alone or the conjugate TFO-CPT(10) or the control oligonucleotide TFO-NPh2. The samples were lysed with an ionic detergent and proteins that were covalently attached to genomic DNA were separated from free proteins by sedimentation through a CsCl gradient by ultracentrifugation. As shown in Fig. 4, 12 fractions were recovered and analyzed by Western slot blotting to detect the fractions containing Topo I. In the control samples with no drug, Topo I was found exclusively at the top of the CsCl gradient as free protein (fractions 1 to 6). In contrast, with drug-treated cells, Topo I was found in the top fractions, as well as in lower fractions 8 to 11 near the bottom of the gradient, where the nucleic acids were localized (as judged from absorbance measurements at 260 nm), corresponding to Topo I/DNA complexes. Such complexes (bands in fractions 8 to 11) were visualized in the presence of CPT, as expected, but also with TFO-CPT(10). In contrast, covalent complex formation was not detected with the conjugate TFO-NPh2, lacking the CPT moiety. These data are consistent with the fact that TFO-CPT conjugates stabilize Topo I/DNA complexes. Although the immunoassay is essentially qualitative, Fig. 4 illustrates that the drug-trapped covalent complexes (evaluated as the amount of Topo I in the DNA fractions normalized to the total amount of Topo I) were somewhat more abundant with CPT than with TFO-CPT(10), consistent with the respective theoretical number of cleavage sites. It is important to note that the TFO used in this study recognizes in the entire human genome ca. 1,500 sites (of which only 436 are found in genes). This high frequency could explain why we are able to detect some cleavage complexes in the presence of TFO-CPT(10).
FIG. 4.
Immunoblot analysis of Topo I-DNA covalent complexes stabilized by Topo I inhibitor in HeLa cell nuclei. HeLa cell nuclei (5 × 106) were incubated for 3 h at 37°C in the presence or absence of compounds (5 μM). The lysates obtained were applied to a CsCl gradient and centrifuged overnight. Twelve fractions were collected and analyzed by slot blot assay. Fractions 1 to 4 contained free Topo I. Topo I/DNA covalent complexes were detected in fractions 8 to 11. Results are representative of three independent experiments.
Triplex formation associated with a proximal drug consensus site is necessary for Topo I targeting by TFO-CPT conjugates.
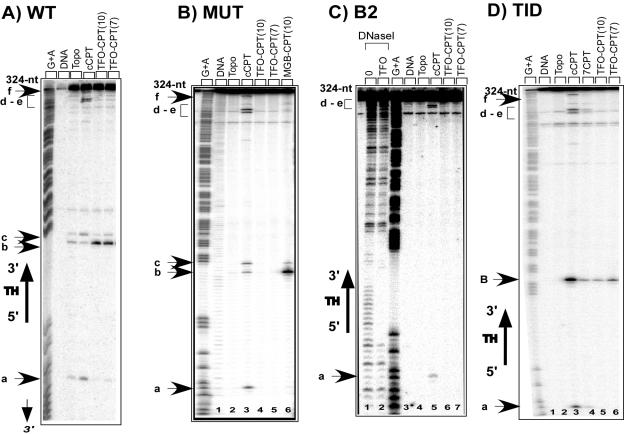
An important issue to address was then the requirements for cleavage complex formation in the presence of the TFO-CPT conjugates. We have previously shown that the linker arm influences the efficacy of cleavage (3, 7) and that the CPT derivatives must be located on the 3′ side of the triplex to induce cleavage (2). Therefore, here we go further by characterizing the molecular specificity of the TFO-CPT conjugates: we evaluated the respective roles of the TFO and of the CPT derivative in the targeted cleavage. To answer these questions, we constructed a series of target duplexes. A 77-bp fragment was inserted between the EcoRI and BamHI sites of plasmid pBSK(+/−) as previously described (7). Four duplex sequences were thus inserted (sequences in Fig. 5): the WT duplex target and three modified ones. The MUT target duplex contains three mutated base pairs (in bold) in the triplex site so that the TFO can no longer bind. The B2 target is modified on the 3′ side of the triplex (in bold) in order to abolish Topo I cleavage sites b and c. Duplex TID was obtained by inserting at the 3′ end of the triplex an 8-bp sequence encompassing a well-known CPT cleavage site from the rRNA gene of Tetrahymena thermophilus characterized by Westergaard and coworkers (1, 25). For all four duplexes, a 324-bp restriction fragment was 32P radiolabeled and incubated in the presence of the conjugates and Topo I. After protease treatment, DNA cleavage products were analyzed by denaturing polyacrylamide gel electrophoresis (Fig. 6).
FIG. 6.
Sequence analysis of the Topo I-mediated cleavage products on the original 324-bp target duplexes (WT), the 324-bp duplex mutated at the triplex site (MUT), and two 324-bp duplexes mutated at sites b and c (B2 and TID). (A) WT: target duplex (lane 1) and duplex incubated with Topo I (lane 2) in the presence of 5 μM cCPT (lane 3), 0.5 μM TFO-CPT(10) (lane 4), or 0.5 μM TFO-CPT(7) (lane 5). (B) MUT: duplex mutated at the triplex site (lane 1) and duplex incubated with Topo I (lane 2) in the presence of 5 μM cCPT (lane 3), 0.5 μM TFO-CPT(10) (lane 4), 0.5 μM TFO-CPT(7) (lane 5), or 1 μM MGB-CPT(10) (lane 6). (C) B2: duplex lacking cleavage sites b and c (lane 3), duplex incubated with DNase I in the absence (lane 1) or presence of 0.5 μM TFO (lane 2), and duplex incubated with Topo I (lane 4) in the presence of 5 μM cCPT (lane 5), 0.5 μM TFO-CPT(10) (lanes 6), or 0.5 μM TFO-CPT(7) (lane 7). (D) TID: duplex containing a known CPT cleavage site, B (lane 1), and duplex incubated with Topo I (lane 2) in the presence of 5 μM cCPT (lane 3) or 7CPT (lane 4), 0.5 μM TFO-CPT(10) (lanes 5), or 0.5 μM TFO-CPT(7) (lane 6). Adenine/guanine-specific Maxam-Gilbert chemical cleavage reactions were used as markers. The positions of the cleavage sites are indicated (sites a to f). The region corresponding to the triplex site is indicated (TH), as is its orientation. nt, nucleotides.
The first question we addressed was whether a triplex site is necessary for targeted cleavage. Figure 6 compares Topo I-mediated cleavage in the presence of TFO-CPT conjugates on the WT and on the MUT targets, differing by the presence of the triplex site. As expected in the case of the WT duplex, triplex formation with the TFO-CPT(10) conjugate (lane 4) strongly enhanced cleavage only at the closest site, b, on the 3′ side of the triplex target, consistent with sequence-specific recruitment of Topo I by triplex formation, as already described for other types of conjugates, including TFO-CPT(7) (lane 5) (2, 5-7). Free CPT cleavage sites (lane 3) were completely abolished, i.e., site a 8 bp from the 5′ end of the triplex, site c on the 3′ side of the triplex site, and sites d, e, and f farther down. On the mutated 324-mer fragment containing three mutated base pairs (in bold) inside the triple-helix site (5′ GAGACTCAGAAAAAAA 3′/3′ CTCTGAGTCTTTTTTT 5′ MUT), the TFO can no longer bind (data not shown). The 3-bp modification does not qualitatively affect the Topo I DNA cleavage pattern in the presence of 5 μM CPT (Fig. 6, compared MUT lane 3 with WT lane 3). However, on the mutated target duplex, the TFO-CPT conjugates TFO-CPT(10) and TFO-CPT(7) did not induce Topo I-mediated DNA cleavage at site b (MUT lanes 4 and 5) under conditions in which it produced specific Topo I-mediated DNA cleavage on the unmodified target duplex (WT lanes 4 and 5, respectively). As a further control, the cCPT analog was attached to a hexa(N-methylpyrrole)carboxamide that binds as a hairpin in the minor groove to A4/T4 tracts (MGB-cCPT conjugate in Fig. 1B). Its binding site is still present in the mutated duplex target MUT, and therefore it induced Topo I-mediated DNA cleavage specifically in the vicinity of its binding site, sites b and c (lane 6 of MUT). These results demonstrate that Topo I-mediated DNA cleavage by the conjugates is based on triplex formation, and when the triple-helical structure cannot be formed on a mutated target, no cleavage is observed.
The second question we addressed was whether the presence of a drug-specific site in the vicinity of the triple helix is necessary to induce DNA cleavage. Figure 6 compares Topo I-mediated DNA cleavage in the presence of the TFO-CPT conjugates on a 324-mer DNA target modified at sites b and c, at 4 bp and 7 bp, respectively, from the triplex 3′ end (5′ GAGAGAGAGAAAAAAAGAGTTTTTCT 3′/3′ CTCTCTCTCTTTTTTTCTCAAAAAGT 5′ in duplex B2). As expected, this mutation (in bold) affects the Topo I DNA cleavage profile in the presence of 5 μM CPT only at sites b and c, which disappear on the mutated target (compare B2 lane 5 to WT lane 3). The binding of the TFO to its site is not affected, as shown by DNase I footprinting analysis (compare B2 lane 2 to lane 1). When this modified duplex was used, in which sites b and c were no longer present, no site appeared on 324-bp fragment B2 in the presence of either the TFO-CPT(10) (lane 6) or the TFO-CPT(7) (lane 7) conjugate. Therefore, the presence of a poison-inducible cleavage site near the triplex on the 3′ side seems to be another factor necessary for effective Topo I targeting. To further characterize this feature, we used a duplex containing, instead of sites b and c, a strong CPT cleavage site (in bold) 4 bp from the 3′ triplex end (distance appropriate for targeting with the TFO-CPT conjugates studied (2, 7), called site B (5′ GAGAGAGAGAAAAAAACTCCAAGTCT 3′/3′ CTCTCTCTCTTTTTTTGAGGTTCAGA 5′] in duplex TID). This site was built from a CPT-sensitive site present in the rRNA gene previously studied by Westergaard and coworkers (1, 25). The cleavage pattern in the presence of CPT was, compared to the unmutated target duplex (WT), modified only at the 3′ side of the triplex site, where site B appeared, located 4 bp from the triplex end (Fig. 6, TID lane 3). The TFO-CPT(10) and TFO-CPT(7) conjugates directed DNA cleavage specifically at site B on the 3′ side of the triplex (TID lanes 5 and 6), in agreement with a sequence-specific recruitment of Topo I upon triplex formation with TFO-drug conjugates. These results confirm that Topo I cleavage targeting by triplex formation is possible when an appropriate CPT-inducible site is present near the triplex 3′ end.
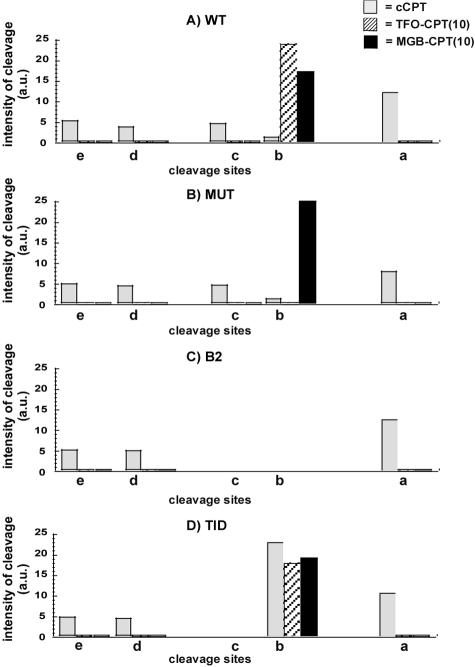
In Fig. 7, the intensities of cleavage, measured as described in Materials and Methods, are reported for each duplex studied at sites a, b, c, d, and e in the presence of free cCPT (gray bars) and conjugates TFO-CPT(10) (hatched bars) and MGB-CPT(10) (black bars), summarizing the results obtained with the mutated duplexes. TFO-CPT(7) behaves the same as TFO-CPT(10) (Fig. 6); therefore, it was omitted for simplification of the figure. Here we demonstrated that the sequence context is essential: mutations either in the triplex site or in the DNA duplex receptor are not tolerated. Finally, it is interesting to see the absence of any CPT-mediated cleavage on the 324-bp mutated DNA fragments (either MUT or B2) in the presence of the TFO-CPT conjugates; the targeted cleavage was abolished as well as the other cleavage sites observed with the free CPT. This is likely due to the polyanionic nature of the TFO-CPT conjugate, which induces repulsion from the DNA double helix and prevents any interaction of the CPT with the Topo I-DNA complex, except for the binding site of the TFO, thus increasing the specificity of Topo I cleavage. These in vitro results support our data obtained with cell nuclei (ICE bioassay), which showed a decrease in the amount of Topo I/DNA complexes with the TFO-CPT compared to free CPT.
FIG. 7.
Quantification of the intensity of cleavage for each target in the presence of CPT (gray bars) at 5 μM, conjugate TFO-CPT(10) (hatched bars) at 0.5 μM, and MGB-CPT(10) (black bars) at 1 μM as described in Materials and Methods. Four independent experiments were analyzed. TFO-CPT(7) behaves the same as TFO-CPT(10), and its data were omitted for simplicity. a.u., arbitrary units.
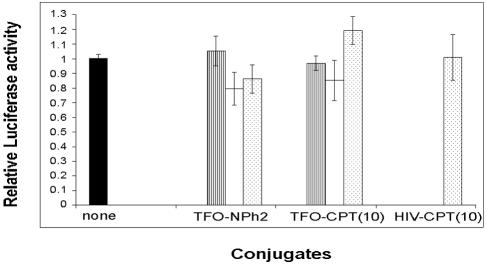
To further validate the importance of the DNA cleavage induced by the TFO-CPT conjugate in cells, we used the fact that if the cleavage sites are mutated, the cleavage activity of TFO-CPTs is abolished. The B2 duplex target modified at its 3′ end by deleting cleavage sites b and c was inserted into firefly luciferase vector pGL3Pr to give pB2, which was cotransfected into HeLa cells with the Renilla luciferase vector and the oligonucleotides at different concentrations (Fig. 8). When the Topo I cleavage site near the 3′ end of the triplex site is deleted, no inhibition is observed in the presence of TFO-CPT(10), as in the case of the control, TFO-NPh2; both are unable to stimulate DNA cleavage. So no inhibition of the luciferase signal is observed when TFO-CPT can no longer stimulate Topo I-mediated DNA cleavage (Fig. 6C and 7C).
FIG. 8.
Cellular activity of the conjugates on pB2 at 24 h after transfection. The firefly luciferase construct was introduced with the control pTK-RL vector in the absence (black bars) or presence of TFO-NPh2, TFO-CPT(10), or HIV-CPT(10) at 0.05 μM (hatched bars), 0.1 μM (white bars), or 0.5 μM (dotted bars). The relative luciferase activity reported is normalized to that of cells in the absence of oligonucleotides (none). The experiments were run in triplicate and repeated 3 to 10 times, and the error bars show standard error calculations. Conditions were the same as in Fig. 2.
DISCUSSION
Recruitment in vitro of Topo I by a TFO linked to a Topo I poison can be achieved with CPT derivatives (6, 19). Here we showed that TFO-CPT conjugates have cellular activity and induce stable cleavage complexes in cell nuclei. In a transient-transfection assay, a TFO-CPT directed specifically against a triplex site situated in the transcribed region of the P. pyralis luciferase reporter gene was able to inhibit 50% of the luciferase activity at 0.5 μM. By contrast, under the same conditions, the untethered TFO and a nonspecific CPT conjugate had no effect. On the other hand, if no cleavage site is available near the triplex end (pB2 construct), no inhibition of the luciferase activity by the TFO-CPT conjugate is observed, supporting evidence that Topo I-mediated DNA cleavage is necessary. In addition, the conjugate induced the formation of covalent Topo I/DNA complexes as visualized by the ICE bioassay. This is the first time that the cellular activity of these conjugates that induce in vitro sequence-specific DNA cleavage by Topo I was measured in cells. This situation differs from that recently reported with Topo II poisons. In fact, daunomycin-TFO conjugates have been shown to inhibit gene expression in cells specifically; however, the inhibition is due to the triplex stabilization effect of the intercalating compound daunomycin and no sequence-specific recruitment of Topo II and consequent DNA cleavage has been shown (12). The better behavior for cleavage of the TFO-CPT conjugates may be explained by the optimized design or by a better adaptability of Topo I versus Topo II, which is considerably bigger than Topo I.
Concerning the efficacy of the TFO-CPT conjugate (50% reduction of the luciferase signal at 0.5 μM), it is comparable to that found in other studies with luciferase reporter systems and pyrimidine or purine triplexes (12, 13, 15; see reference 10 for a review).
Moreover, we further defined the activity of the conjugates and investigated their molecular specificity. By using a series of mutated target sequences, we showed that cleavage site selection is clearly due to triple-helix formation with the conjugate and also to the presence of a pre-existing CPT-sensitive site in close proximity to the triplex end, where the Topo I poison is positioned. As represented in Fig. 6, on the original target (WT), two of the multiple CPT-sensitive sites, b and c, are present 4 bp and 7 bp, respectively, from the 3′ end of the triplex site. When the triple helix is formed with the 16-mer TFO-CPT conjugates, cleavage is strongly enhanced at the site closest to the 3′ end of the triplex (site b) and disappears at all of the other sites. No cleavage is observed on the entire fragment in two situations, (i) when the target is mutated at the triplex site (MUT) and no triple helix can be formed by the conjugate (Fig. 6B) and (ii) if CPT-induced cleavage sites are not present on the 3′ side of the triplex (B2) and no recruitment of Topo I-mediated DNA cleavage is possible (Fig. 6C). Finally, if a strong cleavage site (site B) is created on the 3′ side of the triplex (TID), the triple helix formed with the TFO-CPT conjugate induces specific cleavage at this site (Fig. 6D).
In summary, binding of the TFO to this DNA site positions the Topo I poison such that it inhibits the religation of the Topo I cleavage complex, provided that a Topo I cleavage site is located at the appropriate position with respect to the triple-helical structure, as observed here in in vitro and cellular experiments. Therefore, mutations either in the triplex site or in the DNA duplex receptor are not tolerated; the TFO-CPT conjugate behaves as a Topo I poison with increased specificity compared to the free CPT, as supported by our results obtained in vitro and with cells. Indeed, the negatively charged tail eliminates TFO binding to DNA or DNA/Topo I sites (due to electrostatic repulsion) except at the site where the TFO and the CPT simultaneously find an appropriate target sequence. These rules complete the panel of requirements for optimal triplex-directed CPT activity previously described, which are that the orientation of the conjugate in the cleavage complex is important for the cleavage activity (2) and that the length of the linker arm and the point of attachment on the poison must be optimized within the conjugate (7).
In conclusion, we are now able to control the design of the best TFO-CPT conjugate for Topo I-mediated DNA cleavage recruitment at a specific site. Moreover, we have shown that the conjugates can function as regulators of gene expression in cells. It is also important to remember that long infusion times with CPT are necessary to compensate for the reversibility of the cleavage complexes to achieve maximal activity and that the most effective CPT derivatives form the most stable cleavage complexes (26). In the case of the TFO conjugates, reversibility is greatly reduced (7). Therefore, in the future, triple-helix-directed targeting of antitumor-active Topo I poisons, such as CPT analogs, may be exploited further to improve the efficacy of chemotherapeutic cancer treatments by reducing drug toxicity and targeting Topo I-induced cleavage to appropriately chosen genes. With the appropriate oligonucleotide sequence design, genes involved in the tumor-linked processes can be simultaneously targeted by TFO-drug conjugates.
Supplementary Material
Acknowledgments
We thank Laurent Lacroix and Jean-Louis Mergny for helpful discussions and Erika Brunet for technical help.
This work was supported by grants from the Ligue Nationale Contre le Cancer and ACI “Molécules et Cibles thérapeutiques” of the Ministère de la Recherche, to D.G. from the Fondation de France (Comité Leucémie), and to K.O. from the Assocation pour la Recherche sur les Tumeurs de la Prostate. This work was supported at the University of Virginia by NIH research grant CA78415, awarded by the National Cancer Institute.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andersen, A. H., E. Gocke, B. J. Bonven, O. F. Nielsen, and O. Westergaard. 1985. Topoisomerase I has a strong binding preference for a conserved hexadecameric sequence in the promoter region of the rRNA gene from Tetrahymena pyriformis. Nucleic Acids Res. 13:1543-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arimondo, P. B., S. Angenault, L. Halby, A. Boutorine, F. Schmidt, C. Monneret, T. Garestier, J. S. Sun, C. Bailly, and C. Helene. 2003. Spatial organization of topoisomerase I-mediated DNA cleavage induced by camptothecin-oligonucleotide conjugates. Nucleic Acids Res. 31:4031-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arimondo, P. B., C. Bailly, A. Boutorine, P. Moreau, M. Prudhomme, J. S. Sun, T. Garestier, and C. Helene. 2001. Triple helix-forming oligonucleotides conjugated to indolocarbazole poisons direct topoisomerase I-mediated DNA cleavage to a specific site. Bioconjug. Chem. 12:501-509. [DOI] [PubMed] [Google Scholar]
- 4.Arimondo, P. B., C. Bailly, A. Boutorine, M. Prudhomme, J.-S. Sun, T. Garestier, and C. Hélène. 2000. Recognition and cleavage of DNA by rebeccamycin- or benzopyridoquinoxaline-triplex-forming oligonucleotide conjugates. Bioorg. Med. Chem. 8:1-8. [DOI] [PubMed] [Google Scholar]
- 5.Arimondo, P. B., C. Bailly, A. Boutorine, V. Ryabinin, A. Syniakov, J. S. Sun, T. Garestier, and C. Helene. 2001. Directing topoisomerase I-mediated DNA cleavage to specific sites by camptothecin tethered to minor and major groove ligands. Angew. Chem. Int. Ed. Engl. 40:3045-3048. [DOI] [PubMed] [Google Scholar]
- 6.Arimondo, P. B., C. Bailly, A. Boutorine, J. S. Sun, T. Garestier, and C. Hélène. 1999. Targeting topoisomerase I cleavage to specific sequences of DNA by triple helix-forming oligonucleotide conjugates. A comparison between a rebeccamycin derivative and camptothecin. C. R. Acad. Sci. Ser. III Life Sci. 322:785-790. [DOI] [PubMed] [Google Scholar]
- 7.Arimondo, P. B., A. Boutorine, B. Baldeyrou, C. Bailly, M. Kuwahara, S. M. Hecht, J. S. Sun, T. Garestier, and C. Helene. 2002. Design and optimization of camptothecin conjugates of triple helix-forming oligonucleotides for sequence-specific DNA cleavage by topoisomerase I. J. Biol. Chem. 277:3132-3140. [DOI] [PubMed] [Google Scholar]
- 8.Bailly, C., C. OhUigin, C. Rivalle, E. Bisagni, J. P. Henichart, and M. J. Waring. 1990. Sequence-selective binding of an ellipticine derivative to DNA. Nucleic Acids Res. 18:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailly, C., J. F. Riou, C. Houssier, E. Rodrigues-Pereira, and M. Prudhomme. 1997. DNA cleavage by topoisomerase I on the presence of indolocarbazole derivatives of rebeccamycin. Biochemistry 36:3917-3929. [DOI] [PubMed] [Google Scholar]
- 10.Besch, R., C. Giovannangeli, and K. Degitz. 2004. Triplex-forming oligonucleotides—sequence-specific DNA ligands as tools for gene inhibition and for modulation of DNA-associated functions. Curr. Drug Targets 5:691-703. [DOI] [PubMed] [Google Scholar]
- 11.Cantor, C. R., M. M. Warshaw, and H. Shapiro. 1970. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers 9:1059-1077. [DOI] [PubMed] [Google Scholar]
- 12.Carbone, G. M., E. McGuffie, S. Napoli, C. E. Flanagan, C. Dembech, U. Negri, F. Arcamone, M. L. Capobianco, and C. V. Catapano. 2004. DNA binding and antigene activity of a daunomycin-conjugated triplex-forming oligonucleotide targeting the P2 promoter of the human c-myc gene. Nucleic Acids Res. 32:2396-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carbone, G. M., S. Napoli, A. Valentini, F. Cavalli, D. K. Watson, and C. V. Catapano. 2004. Triplex DNA-mediated downregulation of Ets2 expression results in growth inhibition and apoptosis in human prostate cancer cells. Nucleic Acids Res. 32:4358-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Champoux, J. J. 2001. DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 15.Faria, M., C. D. Wood, L. Perrouault, J. S. Nelson, A. Winter, M. R. White, C. Helene, and C. Giovannangeli. 2000. Targeted inhibition of transcription elongation in cells mediated by triplex-forming oligonucleotides. Proc. Natl. Acad. Sci. USA 97:3862-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Froehler, B. C., R. J. Jones, X. D. Cao, and T. J. Terhorst. 1993. Oligonucleotides derived from 5-(1-propynyl)-2′-O-allyl-uridine and 5-(1-propynyl)-2′-O-allyl-cytidine—synthesis and RNA duplex formation. Tetrahedron Lett. 34:1003-1006. [Google Scholar]
- 17.Kim, D. K., and N. Lee. 2002. Recent advances in topoisomerase I-targeting agents, camptothecin analogues. Mini Rev. Med. Chem. 2:611-619. [DOI] [PubMed] [Google Scholar]
- 18.Lacroix, L., J. Lacoste, J. F. Reddoch, J. L. Mergny, D. D. Levy, M. M. Seidman, M. D. Matteucci, and P. M. Glazer. 1999. Triplex formation by oligonucleotides containing 5-(1-propynyl)-2′-deoxyuridine: decreased magnesium dependence and improved intracellular gene targeting. Biochemistry 38:1893-1901. [DOI] [PubMed] [Google Scholar]
- 19.Matteucci, M., K.-Y. Lin, T. Huang, R. Wagner, D. D. Sternbach, M. Mehrotra, and J. M. Besterman. 1997. Sequence-specific targeting of duplex DNA using a camptothecin-triple helix forming oligonucleotide conjugate and topoisomerase I. J. Am. Chem. Soc. 119:6939-6940. [Google Scholar]
- 20.Pommier, Y., P. Pourquier, Y. Fan, and D. Strumberg. 1998. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta 1400:83-105. [DOI] [PubMed] [Google Scholar]
- 21.Pommier, Y., P. Pourquier, Y. Urasaki, J. Wu, and G. S. Laco. 1999. Topoisomerase I inhibitors: selectivity and cellular resistance. Drug Resist. Updates 2:307-318. [DOI] [PubMed] [Google Scholar]
- 22.Pommier, Y., C. Redon, V. A. Rao, J. A. Seiler, O. Sordet, H. Takemura, S. Antony, L. Meng, Z. Liao, G. Kohlhagen, H. Zhang, and K. W. Kohn. 2003. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 532:173-203. [DOI] [PubMed] [Google Scholar]
- 23.Prudhomme, M. 2000. Recent developments of rebeccamycin analogues as topoisomerase I inhibitors and antitumor agents. Curr. Med. Chem. 7:1189-1212. [DOI] [PubMed] [Google Scholar]
- 24.Thomas, C. J., N. J. Rahier, and S. M. Hecht. 2004. Camptothecin: current perspectives. Bioorg. Med. Chem. 12:1585-1604. [DOI] [PubMed] [Google Scholar]
- 25.Thomsen, B., S. Mollerup, B. J. Bonven, R. Frank, H. Blocker, O. F. Nielsen, and O. Westergaard. 1987. Sequence specificity of DNA topoisomerase I in the presence and absence of camptothecin. EMBO J. 6:1817-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wadkins, R. M., D. Bearss, G. Manikumar, M. C. Wani, M. E. Wall, and D. D. Von Hoff. 2004. Topoisomerase I-DNA complex stability induced by camptothecins and its role in drug activity. Curr. Med. Chem. Anti-cancer Agents 4:327-334. [DOI] [PubMed] [Google Scholar]
- 27.Wang, C. C., and P. B. Dervan. 2001. Sequence-specific trapping of topoisomerase I by DNA binding polyamide-camptothecin conjugates. J. Am. Chem. Soc. 123:8657-8661. [DOI] [PubMed] [Google Scholar]
- 28.Wang, J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3:430-440. [DOI] [PubMed] [Google Scholar]
- 29.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]
- 30.Woo, M. H., J. R. Vance, A. R. Marcos, C. Bailly, and M. A. Bjornsti. 2002. Active site mutations in DNA topoisomerase I distinguish the cytotoxic activities of camptothecin and the indolocarbazole, rebeccamycin. J. Biol. Chem. 277:3813-3822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.