Abstract
The multiple isoforms of the transmembrane glycoprotein CD44 are produced by alternative RNA splicing. Expression of CD44 isoforms containing variable 5 exon (v5) correlates with enhanced malignancy and invasiveness of some tumors. Here we demonstrate that SRm160, a splicing coactivator, regulates CD44 alternative splicing in a Ras-dependent manner. Overexpression of SRm160 stimulates inclusion of CD44 v5 when Ras is activated. Conversely, small interfering RNA (siRNA)-mediated silencing of SRm160 significantly reduces v5 inclusion. Immunoprecipitation shows association of SRm160 with Sam68, a protein that also stimulates v5 inclusion in a Ras-dependent manner, suggesting that these two proteins interact to regulate CD44 splicing. Importantly, siRNA-mediated depletion of CD44 v5 decreases tumor cell invasion. Reduction of SRm160 by siRNA transfection downregulates the endogenous levels of CD44 isoforms, including v5, and correlates with a decrease in tumor cell invasiveness.
CD44 is a transmembrane glycoprotein that mediates the response of cells to their cellular microenvironment. CD44 is expressed in most tissues, where its gene products function in lymphocyte homing, adhesion, migration, and regulation of cell growth (for review, see reference 35). This variety of roles results from multiple CD44 isoforms produced by alternative splicing (3). The CD44 gene is composed of 10 constitutively spliced exons and 10 variable exons, residing between constitutive exons 5 and 6.
Interestingly, the peptides encoded by these variable exons are located in the extracellular domain of the protein. Some of these alternative spliced variants interact with growth factors and render cells responsive to extracellular stimuli (2, 17, 33, 37-39, 43). The CD44 splice variant containing variable exon 6 (v6) can form a complex with the extracellular hepatocyte growth factor (HGF) and its tyrosine kinase receptor Met (33). Formation of this CD44 v6-HGF-Met complex stimulates the activation of Met through autophosphorylation and further activates Met-dependent Ras signaling, probably through the association of ERM (ezrin-radixin-moesin) proteins to the cytoplasmic tail of CD44 (33). The net result is that the CD44 v6-HGF-Met complex activates Ras signaling and promotes cell proliferation. Interestingly, Ras activation also stimulates transcription of CD44 and promotes inclusion of its variable exons (14, 19). This constitutes a CD44-mediated positive feedback loop in the activation of Ras signaling if a factor such as HGF is present. Alternatively, the tumor suppressor protein merlin can also bind to the cytoplasmic tail of CD44. Binding of merlin to CD44 disrupts the interaction between ERM and CD44 and therefore inhibits Ras activation (29). As a result, Ras-dependent CD44 alternative splicing is inhibited, the positive feedback loop is disrupted, and cell growth diminishes. Consistent with the above observations, the majority of CD44 expressed in resting lymphocytes is in the constitutively spliced form (standard form), whereas the variants are commonly observed only during activation.
When overexpressed in human colon cancer cells, the standard form of CD44 acts as a tumor suppressor and inhibits the potential for metastasis (5, 34). Furthermore, a significant body of evidence indicates a correlation between metastasis and dysregulation of CD44 variants. For example, ectopically expressed CD44 v4-7 variant (containing variable exons 4, 5, 6, and 7) confers metastatic potential to nonmetastatic cells (11). Conversely, high levels of expression of CD44 variants correlate with tumors with a poor clinical prognosis (12, 13, 20, 30, 31, 46). Despite the significance of the association of CD44 variants with a variety of malignancies, insight into the regulation of CD44 alternative splicing remains incomplete.
Regulation of CD44 alternative splicing has been studied following mitogen stimulation. Using a splicing construct containing the CD44 v5 exon flanked by its introns and two constitutive exons, Konig and colleagues found that inclusion of v5 is activated by the Ras-Raf-MEK-Erk (extracellular signal-regulated kinase) signaling cascade (23, 45). Addition of the phorbol ester 12-o-tetradecanoylphorbol-13-acetate (PMA), a Ras signaling activator, or ectopic expression of the above signaling molecules elevates the inclusion of v5. Analysis of cis-acting elements in the 117-nucleotide (nt) v5 exon revealed the presence of both exonic splicing enhancers (ESEs) and exonic splicing silencers. These cis-acting elements were identified by consecutively substituting 10-nt mutations through the v5 region (19). Interestingly, in contrast to the broad distribution of experimentally identified exonic splicing silencers, the v5 ESEs are located in two 10-nt segments. The most defective mutant was created by the substitution of the v5 ATGAAGAGGA sequence with random sequence. Mutational substitution of this region reduced basal level inclusion of the v5 exon and decreased the response to PMA stimulation. The proteins that specifically interact with this ESE in this context have yet to be identified. Recently, a nuclear RNA binding protein, Sam68, was shown to bind to a region of v5 containing the site AAAAUU (21, 23). The activation of Ras signaling by PMA, which phosphorylates and in turn activates Sam68, stimulates v5 exon inclusion (23). Deletion of the Sam68 binding site does not inactivate the PMA response, consistent with the notion that other factors in addition to Sam68 are also important.
In this study, we show that SRm160, a splicing activator of pre-mRNAs containing GAA repeats (4, 8), specifically stimulates inclusion of CD44 v5 in a Ras-dependent manner. Mutation of the above ATGAAGAGGA sequence abolishes SRm160's stimulation. SRm160 and Sam68 are detected in a complex that probably regulates CD44 alternative splicing. Moreover, we show that small interfering RNA (siRNA) knockdown of CD44 v5 decreases tumor cell invasion. Reduction of SRm160 inhibits alternative splicing of endogenous CD44 variants and results in a decrease in tumor cell invasiveness. These observations support a link between regulation of alternative splicing and tumor cell invasiveness.
MATERIALS AND METHODS
Cells and transfection.
Human HeLa cells and 293T cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, penicillin, and streptomycin. For transfection of 293T cells, cells were plated in 24-well plates 1 day before transfection. Cells were transfected with 0.8 μg of plasmids (including v5 Photinus luciferase reporter and Renilla luciferase as an internal transfection control) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. At 24 h after transfection, cells were collected. One-fifth of the cells was used for RNA analysis (see below). The remaining four-fifths were lysed in 100 μl of passive lysis buffer. Luciferase activity was assayed using a dual-luciferase kit (Promega).
RNA interference (RNAi) experiments were performed in HeLa cells using 100 pmol of siRNAs (control, SRm160, Sam68, or CD44 v5) and 5 μl of oligofectamine for each well of a six-well plate. For cotransfection with the CD44 v5 reporter gene, siRNA-treated HeLa cells were trypsinized and plated in 12-well plates 24 h after the first siRNA transfection. Twenty-four hours after the cells were plated, the CD44 reporter construct was cotransfected with an H-Ras V12 plasmid and the same siRNAs transfected in six-well plates using Lipofectamine 2000. Cells were harvested 30 to 36 h posttransfection. siRNA sequences of the sense strands are as follows: SRm160-1, CGACCCAAGAGAUCCCAUGdTdG; SRm160-2, GAGACGUUCACCUUCAUUAdTdT; Sam68-1, CGGAUAUGAUGGAUGAUAUdTdT; Sam68-2, CGCAGAACAAAGUUACGAAdTdT; CD44 v5-1, CCACUGCUUAUGAAGGAAAdTdT; and CD44v5-2, AUGAGCAUCAUGAGGAAdTdT.
RT-PCR.
RNA was prepared using a QIAGEN RNeasy kit. A total of 0.25 μg of RNA was used in a reverse transcription (RT) reaction in a 20-μl volume for 1 h. Two microliters of the resulting cDNA was subjected to PCRs using specific primers. Each cycle of PCRs included 45 s at 94°C, 45 s at 55°C, and 45 s at 72°C. Thirty cycles were performed to detect v5 inclusion using the v5 luciferase minigene construct. For endogenous CD44, different cycles of PCR were performed, as follows: 30 cycles for v8-10; 31 cycles for v3r; 32 cycles for v3, v7, v6r, and v7r; 34 cycles for v2r; and 35 cycles for v2, v4, v5, and v6. Twenty-nine cycles were used for amplifying standard CD44 and loading control dihydrofolate reductase (DHFR). PCR products were analyzed on a 1.5% agarose gel. SRm160 was amplified using the forward primer 5′-GTCGTGTTTCTGTGTCTCCAG and the reverse primer 5′-TATGCTTGGATGATAATGAAGGTG. DHFR was amplified using the forward primer 5′-GTTCTGCTGTAACGAGCGG and the reverse primer 5′-AGGCATCATCTAGACTTCTGG. The primers for amplifying the CD44 luciferase minigene and endogenous CD44 variants using forward individual variable exon primers were described previously (18, 45). A forward primer pairing to the 5′ constitutive region is 5′-CATCCCAGACGAAGACAGTC. Reverse primers base pairing to specific variable exons are as follows: v2r, 5′-TGTGAAGATGATTCTTTGACTC; v3r, 5′-CATCATCAATGCCTGATCCAGA; v6r, 5′-CAGCTGTCCCTGTTGTCGAA; and v7r, 5′-TCCTGCTTGATGACCTCGTC.
Immunoprecipitation.
293T cells transfected with hemagglutinin (HA)-tagged Sam68 or Flag-tagged SRm160 were lysed in NP-40 lysis buffer (150 mM NaCl, 50 mM Tris, 1% NP-40, 50 mM NaF, supplemented with protease inhibitors [complete tablet from Roche]). Cell lysates were precleared with protein A beads and incubated with HA or Flag antibody-conjugated beads (Roche and Sigma, respectively). The immunoprecipitated complexes were washed five times with NP-40 buffer and eluted in 2× sodium dodecyl sulfate (SDS) loading buffer by incubation at 95°C for 5 min. The eluates were loaded onto an SDS-polyacrylamide gel for Western blot analysis.
Matrigel invasion assay.
Biocoat Matrigel invasion chambers (Becton Dickinson) were used to assess the invasiveness of HeLa cells. Two days after siRNA treatment, 5 × 104 cells were resuspended in 250 μl of serum-free Dulbecco's modified Eagle's medium and added to the cell culture inserts of the invasion chambers. Fetal bovine serum (10%) was used as a chemoattractant and placed in the lower wells. After 22 to 24 h, cells on the upper surface of the membrane were removed using cotton swabs, and the filters were fixed by immersion in 4% formaldehyde for 10 min. After two washes with water, the invaded cells were then stained with 0.2% crystal violet. Excess dye was rinsed off two times with water. The stained cells were photographed using a digital camera and counted.
RESULTS
Overexpression of SRm160 stimulates CD44 v5 inclusion dependent on Ras signaling.
Because of the biological importance of CD44 splice variants in tumorigenesis, we explored the regulation of CD44 alternative splicing. The CD44 variable exon sequences were compared between species, and potential ESEs in the v5 exon were computationally identified (9). Among the species examined (human, rat, and mouse), conserved GAA repeats were observed in the v5 exon. Both experimental and computational analyses indicate that GAA repeats are a hallmark of ESEs that activate splicing (9, 40). Interestingly, SRm160, an SR-containing splicing coactivator, has been shown to be important in the splicing of pre-mRNAs that contain GAA repeats (8). These observations suggested that SRm160 might be involved in the regulation of v5 inclusion.
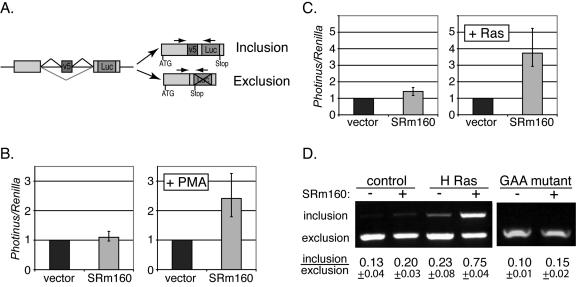
To determine whether SRm160 has an important role in regulating CD44 alternative splicing, we used a CD44 v5 reporter minigene construct (23, 45). This minigene contains a luciferase-based splicing reporter, in which the CD44 v5 exon and its flanking introns were inserted upstream of an intact Photinus luciferase gene (Fig. 1A). Inclusion of the v5 exon produces a fusion protein containing the coding sequence of the Photinus luciferase, whereas exclusion of the v5 exon positions a stop codon upstream of the luciferase coding region. Thus, the luciferase activity directly indicates the levels of CD44 v5 inclusion. Previous work has demonstrated that the ratios of the v5 inclusion and exclusion are consistent whether they are measured by RT-PCR of spliced mRNA or luciferase assays (45).
FIG. 1.
SRm160 stimulates CD44 v5 inclusion. (A) The luciferase-based v5 minigene schematic. Exons are shown as boxes and introns are denoted as straight lines. Lines above and below each intron illustrate different joining of exons by alternative splicing. Positions of the translation start codon (ATG) and stop codon are shown. Inclusion of v5 results in expression of an in-frame luciferase protein, whereas exclusion of v5 yields a stop codon upstream of the luciferase gene. The relative locations of the primers used in the PCRs to detect the included and excluded forms are shown as arrows at the top of these forms. (B) Dual luciferase assay of 293T cells. Cells were transiently transfected with the v5 minigene (Photinus luciferase), a Renilla luciferase plasmid (used as an internal transfection control), and either an empty vector (black bars) or SRm160 expression plasmid (gray bars). About 18 h after transfection, cells were treated without PMA (left panel) or with PMA (right panel) for 6 h. After this treatment, cells were harvested and lysed. Luciferase activities were assayed using a Dual-Glo luciferase kit (Promega). The ratios of Photinus and Renilla luciferase activities were normalized to the empty vector transfections (black bars). Error bars denote the range of experimental variations. (C) Relative luciferase activity of cells transiently transfected with the v5 minigene plus either empty vector (black bars) or SRm160 plasmid (gray bars) and cotransfected without or with activated H-Ras V12 plasmid. The ratios of Photinus and Renilla luciferase activities were normalized to the non-SRm160 (empty vector) transfections. (D) RT-PCR analysis of harvested RNA from cells transfected with the constructs described in panel B (without or with SRm160 and without or with activated H-Ras V12) or with a GAA-mutant v5 construct (in the presence of activated H-Ras V12, without or with SRm160). Relative locations of the PCR primers are shown in panel A. PCR products were analyzed in 1.5% agarose gels. The inclusion and exclusion forms of v5 were detected in one PCR. Positions of these two forms are depicted on the left of the gel. Calculated ratios between these forms are shown at the bottom of the gel.
The CD44 v5 minigene construct was cotransfected in 293T cells with either a control vector or a plasmid ectopically expressing SRm160, and a Renilla luciferase plasmid served as an internal transfection control. One transfection (Fig. 1B, vector) included the CD44 v5 minigene and an empty plasmid vector. The second transfection (Fig. 1B, SRm160) included the v5 minigene and the plasmid encoding SRm160. This series was repeated with the addition of treatment with PMA, which activates the Ras pathway. The level of luciferase was normalized to 1.0 following transfection with the control plasmid, which corrects for any stimulation by PMA alone of either transcription or inclusion of v5. Under these conditions, SRm160 overexpression increased the level of luciferase, i.e., CD44 v5 inclusion, by 2.4-fold in comparison with control vector transfection (Fig. 1B). This increase was entirely dependent upon PMA stimulation, as very little change (1.1-fold) was produced by SRm160 overexpression in non-PMA-treated cells. These observations suggested that stimulation of CD44 alternative splicing by SRm160 is regulated by Ras signaling. To confirm this hypothesis, a constitutively activated form of Ras, H-Ras V12, was ectopically expressed in 293T cells along with either control or SRm160 expression plasmids using the same experimental design (Fig. 1C). As expected, SRm160 specifically stimulated luciferase expression by ∼3.7-fold in cells that overexpressed H-Ras V12. These results are consistent with the idea that the stimulation of v5 inclusion by SRm160 is dependent on Ras signaling.
The stimulation of luciferase expression from the CD44 v5 minigene is an indirect measure of alternative splicing. To directly assay the v5 inclusion in mRNA, we analyzed the ratios of variants by RT-PCR with primers flanking the v5 exon (Fig. 1A; the location of primers relative to spliced mRNAs is indicated by arrows). Both the included and excluded variants can be detected in one PCR, with the former yielding a longer product. In the absence of Ras activation, the levels of v5 inclusion were similar in cells ectopically overexpressing SRm160 or control vector (0.20 and 0.13, respectively). However, in the presence of cotransfected activated Ras, overexpression of SRm160 stimulated the level of v5 inclusion to 0.75 (Fig. 1D). These results show that SRm160 activity can regulate CD44 alternative splicing through processes influenced by Ras signaling.
A potential mechanism explaining the activity of SRm160 in stimulation of the v5 exon is the presence of GAA-containing ESEs. This was addressed by testing the activity of a mutant form of v5 exon where the 10-nt GAA-containing ESE segment (ATGAAGAGGA) was replaced with a random sequence (19). In agreement with previously described results (19), we observed, in the presence of activated Ras, an ∼50% reduction in inclusion of the v5 exon when this GAA mutant was compared to the wild-type v5 construct. More importantly, the SRm160 stimulatory effect on v5 inclusion was almost entirely abolished by this mutation (0.10 versus 0.15, without and with SRm160, respectively) (Fig. 1D). A control mutation with a 10-nt substitution immediately downstream of this GAA segment displayed stimulatory effects very similar to the wild-type v5 construct upon SRm160 overexpression (ratios of inclusion to exclusion: 0.23 versus 1.09, without and with SRm160, respectively) (data not shown). To test whether the 10-nt GAA-containing ESE segment is sufficient to allow Ras-dependent alternative splicing, we used a reporter construct, pSXN (6, 9), that contains an alternatively spliced 33-nt exon 2. Random-sequence insertion in this exon 2 predominantly results in exon skipping. Interestingly, insertion of the 10-nt GAA segment into the pSXN exon 2 promoted inclusion of this exon. This inclusion was moderately stimulated (1.6-fold ± 0.1-fold) in response to SRm160 in a Ras signaling-dependent manner (data not shown). These observations indicate that the v5 GAA-containing ESEs and possibly other elements are important for SRm160's stimulation of v5 inclusion.
Knockdown of SRm160 inhibits inclusion of the CD44 v5 exon.
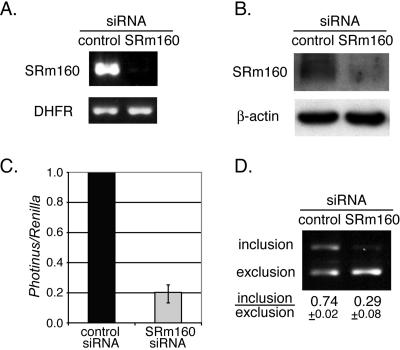
To further investigate the role of SRm160, we determined the effects of SRm160 reduction on v5 inclusion. SRm160 siRNA transfection was used to reduce the level of this protein. Transient transfection of the SRm160 siRNA produced an ∼10-fold knockdown of SRm160 mRNA as measured by RT-PCR analysis relative to cells transfected with a control siRNA (Fig. 2A). Protein analysis using immunoblotting with an SRm160-specific antibody showed that the levels of a series of bands of the SRm160 protein were undetectable in cells treated with SRm160 siRNA (Fig. 2B). These clustered polypeptide bands for SRm160 have been described previously and probably represent different degrees of phosphorylation of this highly repetitive SR protein. In total, these results indicate that RNAi knockdown was successful.
FIG. 2.
SRm160 knockdown inhibits v5 inclusion. (A) Quantitative RT-PCR analysis of the SRm160 mRNA harvested from HeLa cells transiently transfected with either control siRNA or SRm160 siRNA. Primers specific to SRm160 were used. Amplification of DHFR mRNA was used as an internal control. (B) Western blot analysis of SRm160 protein levels in siRNA-transfected cells described in panel A. The β-actin protein levels were used as a loading control. (C) Relative luciferase activities of HeLa cells transiently transfected with the v5-Luc minigene, H-Ras V12 plasmid, and a control siRNA or SRm160 siRNA. Ratios of the Photinus and Renilla luciferase activities were normalized to the control siRNA transfections. (D) RT-PCR analysis of RNA from siRNA-transfected cells described in panel C. PCR primers are shown in Fig. 1A. PCR products were analyzed in 1.5% agarose gels. Inclusion and exclusion forms of v5 were detected in one PCR. Positions of these two forms are indicated on the left of the gel. Calculated ratios between these forms are shown at the bottom of the gel.
The v5-Photinus luciferase reporter construct was cotransfected in HeLa cells with the H-Ras V12 plasmid, the internal control Renilla luciferase plasmid, and either SRm160 siRNA or control siRNA. Again, the level of expression of the v5-Photinus luciferase reporter was normalized to 1.0 for the control siRNA. There was about a fivefold reduction of v5 inclusion in cells treated with the SRm160 siRNA in comparison to those treated with the control siRNA (Fig. 2C). These results were verified by measuring mRNA levels of v5 inclusion versus exclusion by RT-PCR. As anticipated, the inclusion form of v5 was reduced in SRm160 siRNA-treated cells. The ratio of included to excluded forms in SRm160 siRNA-treated cells was reduced to 0.29 from 0.74, the level observed in control siRNA-treated cells (Fig. 2D). To exclude the possibility that the observed effect is due to nonspecific suppression of off-target genes by the SRm160 siRNA, we tested a second siRNA to SRm160. Indeed, expression of this second siRNA also reduced the inclusion of CD44 v5 (data not shown), suggesting that the reduction is specific to SRm160 silencing. These observations indicate that the activity of SRm160 is important for v5 inclusion.
SRm160 interacts with Sam68.
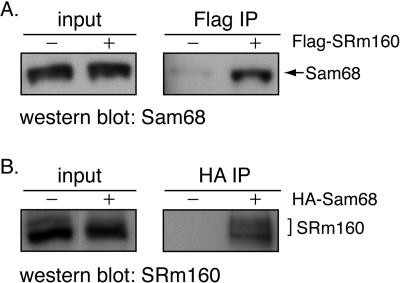
The Src substrate and RNA binding protein Sam68 have been reported to be important for stimulation of v5 inclusion by Ras activation (23). Since both SRm160 and Sam68 stimulate this inclusion, it is possible that these two proteins might act in the same pathway and could interact with each other. To test this possibility, immunoprecipitation experiments were performed. A plasmid containing Flag-tagged SRm160 was transfected into 293T cells. Proteins that interact with Flag-tagged SRm160 protein were precipitated using anti-Flag antibody-coupled beads. The presence of Sam68 in the immunoprecipitated fraction was detected by immunoblotting using a Sam68 antibody. Indeed, the result shows that SRm160 and Sam68 coimmunoprecipitate (Fig. 3A). The above observation was further confirmed by immunoprecipitation using lysates from cells that overexpressed HA-tagged Sam68 protein and anti-HA antibody-coupled beads (Fig. 3B). Again, we observed that SRm160 immunoprecipitated with Sam68. Moreover, this coimmunoprecipitation was resistant to extensive RNase treatment (data not shown), indicating that SRm160 and Sam68 association was not dependent on RNA bridging. We conclude that these two proteins can associate through protein-protein interactions. In addition, we observed that overexpression of Sam68 did not rescue the reduction of v5-luciferase inclusion caused by SRm160 siRNA treatment, suggesting that these two proteins may be functionally cooperative (data not shown).
FIG. 3.
SRm160 and Sam68 coimmunoprecipitate. 293T cells were transfected with plasmids containing either Flag-tagged SRm160 (A) or HA-tagged Sam68 (B) or corresponding empty vectors. Cell lysates were immunoprecipitated using anti-Flag or anti-HA antibodies, respectively (indicated at the top of the gels). Total cell lysates (input) as well as immunoprecipitants (IPs) were analyzed by SDS-polyacrylamide gel electrophoresis. Western blot analyses were performed using antibodies of Sam68 (A, Flag IP) or SRm160 (B, HA IP). The positions of Sam68 and SRm160 are indicated on the right of the gels.
SRm160 regulates splicing of endogenous CD44 variable exons.
The above results, obtained using a transfected plasmid as a reporter for RNA splicing, suggest that SRm160 is important for inclusion of CD44 v5. Therefore, we wanted to determine whether SRm160 regulates the alternative splicing of endogenous CD44. First, the expression pattern of CD44 variants in Ras-activated HeLa cells was examined. As shown in Fig. 4A, a set of 5′ primers that specifically base pair to individual variable exons and a common 3′ constitutive exon were used to detect CD44 variants using quantitative RT-PCR analysis. In this case, the length and sequences of the PCR products from each primer pair should indicate the composition of the CD44 variants containing the specific exon. Using this set of primers, we observed that expression of variants containing v3 to v10 was detectable in HeLa cells. We were not able to assay the levels of v2 since the amplification levels with v2 primers were very low, probably due to the length of the v2 PCR products. In addition, the mobility of the band amplified by the exon 4 primer suggested that the v4 and v5 exons were spliced together (this observation was also verified by sequencing the PCR products). Similarly, exons v7 to v10 are apparently spliced as one unit in these cells. The v6 exon is both joined to the v7 to v10 exons and spliced directly to the 3′ constitutive exon (Fig. 4A).
FIG. 4.
SRm160 is required for splicing of endogenous CD44 variants. (A) Expression pattern of CD44 variants in HeLa cells. The relative locations of PCR primers are depicted at the top of the gels. To analyze different variants of CD44, PCRs were performed using primers that contain different specific 5′ primers pairing to individual variable exons and one 3′ primer pairing to a constitutive exon (filled arrows and indicated as v2 to v10 on top of the gel). PCR amplifications of v2, v3, v6, and v7 variants were also performed using a 5′ primer pairing to a constitutive exon and specific 3′ primers pairing to individual variable exons (open arrows and indicated as v2r, v3r, v6r and v7r). Primers that pair to 5′ and 3′ constitutive exons were used for detection of the CD44 standard form. An agarose gel of PCR products is shown with a 100-bp marker in the left lane. Specific primers used in each PCR are indicated on the top of the gel. (B) RT-PCR analysis of CD44 variant exon inclusion. RNAs were harvested from HeLa cells cotransfected with H-Ras V12 and siRNAs to control, SRm160, and Sam68. Primers for analysis of CD44 variants and the standard form of CD44 are indicated on the left of gels. Amplification of the DHFR mRNA was used as an internal loading control. Agarose gels of PCR analysis of CD44 variants are shown. The relative amounts of inclusion in cells treated with SRm160 and Sam68 siRNAs are normalized to cells treated with control siRNA and plotted as a bar graph. (C) siRNA knockdown of Sam68. RNA analysis of Sam68 from cells treated with control and Sam68 siRNAs is shown in the left panel using Sam68-specific primers. Amplification of the DHFR gene was used as an internal loading control. Western blot analysis of Sam68 knockdown is shown in the right panel. The β-actin protein level was used as a loading control.
Next, the effect of SRm160 knockdown by siRNA was examined. Following SRm160 siRNA treatment, the overall splice patterns of CD44 variants were largely unchanged; however, the levels of expression of most variants were reduced relative to the constitutive isoform (Fig. 4B). For v2, v3, v6, and v7 variants, a second set of PCR primers was used for quantitation. This set contained a 5′ primer complementary to the proximal 5′ constitutive exon and 3′ primers complementary to these individual variable exons (Fig. 4A, v2r, v3r, v6r, and v7r). Each set of primers amplified one major band, corresponding to the joining of the variable exon to the 5′ constitutive exon (v2r, v3r, and v6r) or the joining of both v6 and v7 exons to the constitutive exon (v7r). For quantitation, all of the PCRs were monitored to ensure that measurement was done in a linear range (data not shown). Amplification of the constitutive spliced form of CD44, the product from the two flanking primers, was performed in parallel. Amplification of the DHFR gene was used as an internal control. To compare the expression levels of CD44 between samples (control siRNA- and SRm160 siRNA-treated cells), the CD44 PCR intensities were normalized to those of the DHFR products to obtain relative levels of CD44 expression. Interestingly, we observed that the CD44 constitutive isoform and v10 variant were expressed at similar levels in both groups of siRNA-treated cells. However, expression levels of variants v2 to v9 were reduced in SRm160 siRNA-treated cells. Inclusions of v4 and v5 were most dramatically affected, by five- to sixfold. Inclusion of v3 was decreased by fivefold. Inclusions of v7, v8, and v9 were reduced by three- to fourfold, and v2 and v6 inclusions were decreased by three- and twofold, respectively. As mentioned above, exons v4 and v5 and exons v7, v8, and v9 are spliced as units in HeLa cells (Fig. 4A). The similar degrees of downregulation of these exons provide evidence for the accuracy of the quantitations. Taken together, these results indicate that SRm160 is important for the inclusion of most of the endogenous CD44 variable exons.
Because of the involvement of Sam68 in CD44 alternative splicing and the interaction between Sam68 and SRm160 described above, it was important to determine whether Sam68 is important for inclusion of the same subset of CD44 variable exons as SRm160. To examine this, we knocked down Sam68 expression by siRNAs and examined the effect on CD44 inclusion. RT-PCR analysis showed about a fourfold reduction of Sam68 RNA expression in Sam68 siRNA-treated cells, and Western blot analysis showed a similar reduction of Sam68 protein in these cells (Fig. 4C). Notably, we observed very similar to CD44 inclusion patterns in cells treated with Sam68 siRNA those treated with SRm160 siRNA, with slight differences (Fig. 4B). For example, inclusion of v2 was more affected in Sam68 siRNA- than in SRm160 siRNA-treated cells, with a sevenfold versus a threefold reduction, respectively. Inclusion of v5 was also less affected in the same comparison, with a threefold versus a sixfold reduction, respectively. These results indicate that knockdown of Sam68 impaired the inclusion of endogenous CD44 variable exons to degrees similar to those found with the knockdown of SRm160. Together with the coimmunoprecipitation studies, these results indicate that SRm160 and Sam68 are important for regulation of alternative splicing of the endogenous CD44 gene in HeLa cells.
SRm160 and tumor cell invasion.
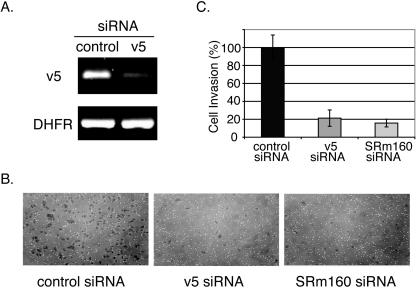
CD44 variants have been implicated in tumor invasion and metastasis (16, 22). Therefore, it is tempting to speculate that the activity of SRm160 could influence tumor cell invasion by regulating the inclusion of CD44 variable exons. Thus, we tested whether reducing SRm160 levels in cells would affect their invasiveness. In order to validate an invasion assay while performing an interesting novel experiment, we tested whether inhibition of CD44 variants by RNAi would affect the invasive properties of cells. We chose to target variants containing v5 exon because v5 inclusion is elevated in some metastatic cells and tissues (12, 13, 20, 46, 47). We used human cervical carcinoma cells (HeLa) as these cells are active in the invasion assay and are highly transfectable (42). An siRNA designed to specifically target the v5 exon was transfected into HeLa cells. As shown in Fig. 5A, we observed a 3.5-fold knockdown of v5-containing variants in cells treated with this siRNA. A control siRNA, whose sequence does not match any known human genes, did not cause a reduction of v5 expression.
FIG. 5.
Knockdown of SRm160 reduces tumor cell invasiveness. (A) Quantitative RT-PCR analysis of the v5 mRNA from HeLa cells transiently transfected with either control siRNA or v5 siRNA. Primers specific to v5 were used. Amplification of DHFR mRNA was used as an internal control. (B) Invasion assays. HeLa cells were transfected with control, CD44 v5, or SRm160 siRNA for 48 h. These cells were then suspended in serum-free medium and placed in an invasion chamber and induced to migrate toward serum-containing medium, requiring invasion through a matrigel-coated membrane. After 22 to 24 h, cells that invaded through the membrane were fixed, stained, and counted. Representative photographs of these cells are shown. (C) Quantitation of tumor cell invasion described in panel B. The percentage of cell invasion was normalized to that of cells transfected with control siRNA.
At 48 h posttransfection, equal numbers of cells treated with either v5 siRNA or control siRNA were added to invasion chambers in the absence of serum. The bottom of the invasion chamber contained a membrane with a layer of reconstituted basement membrane matrix, which impedes noninvasive cells from migrating through the membrane. Invasive cells are able to digest and migrate through the matrix and thus the membrane. Serum-containing medium was placed in the well surrounding the chamber and acted as a chemoattractant for a growth factor-induced invasion. After 22 to 24 h of incubation, we fixed and counted cells that invaded through the matrigel membrane and accumulated in the lower chamber. Interestingly, v5 siRNA-treated cells were impaired in their invasive potential (Fig. 5B and C). The number of v5 siRNA-treated cells passing through the membrane was only approximately 20% of the control siRNA-treated cells. We observed similar results when the above-described experiments were repeated using a second v5 siRNA. These results demonstrate that reduction of the CD44 v5-containing variants inhibits tumor cell invasion. It substantiates the previous suggestion that CD44 variants are involved in cell invasiveness.
Since reduction of SRm160 expression by siRNA knockdown inhibited v5 inclusion, we tested whether treatment with SRm160 siRNA would alter the invasiveness of cells. HeLa cells were treated with siRNAs specific to either SRm160, v5, or nonspecific control siRNA and tested as described above for invasiveness. Interestingly, cells treated with SRm160 siRNA showed a dramatic fivefold reduction in invasion activity (Fig. 5B and C), a reduction in invasiveness similar to that seen with v5 siRNA-treated cells. These results suggested that SRm160 activity could have an important role in tumor cell invasion by altering the level of CD44 variants in cells.
We have previously observed that SRm160 siRNA-treated cells proliferate more slowly than control siRNA-treated cells (unpublished observations). To confirm that the impaired invasiveness by SRm160 was not due to slower cell proliferation, the proliferation rate of these cells was monitored in parallel to the invasion assay. In the invasion assay period, 22 to 24 h, the cell numbers from these two populations differ by less than 15% (data not shown). Therefore, the reduction in cell number in the lower chamber following SRm160 siRNA treatment is not due to the decrease in rate of proliferation.
DISCUSSION
SRm160 regulates CD44 alternative splicing in a Ras-dependent manner.
Many studies have suggested that inclusion of the variable exons of CD44 by alternative splicing correlates with tumor development and metastasis (16, 22, 32, 36). This study indicates that a splicing coactivator, SRm160, can regulate CD44 v5 inclusion in a Ras signaling-dependent manner. We found that ectopically expressed SRm160 stimulated the inclusion of the v5 exon in a transient reporter when the Ras pathway was activated. Furthermore, inclusion of v5 from the same reporter was significantly inhibited when cells were treated with SRm160 siRNA. Previous work has suggested that the splicing factor Sam68 can also regulate the inclusion of the v5 exon in a Ras-dependent manner. We also observed that silencing of Sam68 by siRNA transfection reduced the level of v5 inclusion. This is consistent with the finding that Sam68 and SRm160 can be coimmunoprecipitated. Thus, it is likely that these two splicing factors function in a common pathway in controlling alternative splicing of at least some of the CD44 variable exons.
The ability to silence genes by transfection of mammalian cells with siRNA permits a new test of the roles of particular proteins in control of CD44 alternative splicing. In the case of SRm160, we were able to show that knocking down the level of this protein by more than fivefold after treatment with siRNA resulted in a dramatic reduction in the level of inclusion of several of the alternatively spliced exons expressed from the endogenous CD44 gene. Specifically, inclusion of the v5 exon of CD44 was reduced by sixfold. The standard form of CD44 mRNA, however, was not affected. The activity of SRm160 as a splicing factor for each of the variant exons is unclear. Computational analysis indicates that most of the variable exons could contain ESEs of the GAA-type elements that might recruit SRm160. They also contain AT-rich elements that are potential Sam68 binding sites. It is also possible that the observed reduction in multiple variants after silencing of SRm160 could result from disruption of a positive feedback loop involved in CD44 alternative splicing (our unpublished observations). Treatment of cells with siRNA specific to Sam68 also reduced the levels of inclusion of CD44 variable exons, consistent with the observation that these two splicing factors interact. The similarity of the effects on inclusion of CD44 variable exons when either of these factors was silenced indicates that CD44 alternative splicing is probably regulated by SRm160 and Sam68 through a common pathway.
SRm160 is a coactivator of RNA splicing, which contains many serine and arginine repeats. Some fraction of SRm160 is associated with another highly repetitive protein, SRm300 (4). The GAA-containing ESEs have been shown to be important for recruiting SRm160 to pre-mRNA through interactions with the RNA binding protein Tra2-beta (8, 40). Interestingly, we observed that the function of SRm160 in stimulation of v5 inclusion is dependent on GAA-containing ESEs in this exon. Mutation of this GAA segment abolishes the stimulatory effect of SRm160. In addition, we have observed that SRm160 interacts with another RNA binding protein, Sam68, which has been shown to recognize an AAAUU-type sequence in v5. Since the simultaneous activities of these proteins are necessary for maximal stimulation of v5 inclusion, it is possible that recruitment of SRm160 and Sam68 to v5 pre-mRNA is cooperative. Another indication that Sam68 and SRm160 might have common activity are the recent reports that both factors can stimulate the 3′-end processing of some pre-mRNAs (24, 25, 27).
The mechanism by which Ras activation stimulates CD44 alternative splicing has not been fully determined. Sam68 has been shown to be a target of Erk phosphorylation (23). In addition, the splicing factors, such as SRm160 and Tra2, that contain many phosphorylation sites may also be regulated by Ras signaling. As noted above, SRm160 can associate with SRm300, which is frequently highly phosphorylated in tumor verses normal cells (F. White, personal communication).
v5 inclusion, SRm160, and cell invasion.
The expression of the CD44 v5 variant has been correlated with tumor malignancy in patients with poor clinical prognosis. The prognostic significance of v5 expression has been observed in human thymic epithelial neoplasms and gastric and renal carcinomas (12, 20, 46). Consistent with these clinical observations, our RNAi experiments show that v5 knockdown drastically impairs invasion by tumor cells. It is interesting that siRNA knockdown of v5 in HeLa cells in culture resulted in reduction of other variable exon inclusions (data not shown). This reduction is probably due to inhibition of the positive feedback loop that regulates CD44 alternative splicing (reviewed in the introduction). Thus, it is possible that the decrease in invasiveness of the cells after transfection of the v5 siRNA could also reflect decreases in other variable exons of CD44.
Silencing of SRm160 by siRNA treatment affects the inclusion of CD44 variable exons v2 to v9 without significantly decreasing the level of the constitutive mRNA isoform. The v4 and v5 exon inclusion was the most dramatically affected, with a reduction of five- to sixfold. As might be expected, the decrease in SRm160 causes a parallel decrease in cell invasiveness. Cells treated with SRm160 siRNA had a much lower invasive capacity than cells treated with control siRNA. The phenotype of SRm160 siRNA-treated cells was very similar to that produced by v5 siRNA treatment. This similarity suggests that the primary effect of SRm160 reduction on cell invasion is a consequence of the reduction in v5 levels. However, it is also possible that the reduction of SRm160 by siRNA could also affect invasion through other cellular pathways.
Previous work by others has described interactions between SRm160 and other tumor oncogenes, such as TLS/FUS and DEK (26, 28). TLS/FUS is a nucleic acid-binding protein, and its N-terminal half functions as a transcriptional activator domain in fusion oncoproteins found in human leukemias and liposarcomas (7, 15). Intriguingly, TLS/FUS interacts with SRm160, PTBP, and a subset of the classical SR proteins. DEK is a phosphoprotein that is often fused to the nucleoporin CAN in a subset of acute myeloid leukemias (44). Moreover, as mentioned previously, SRm300, the binding partner of SRm160, was identified as the protein that undergoes the most phosphorylation in cancer cells (F. White, personal communication). Furthermore, Sam68, another protein to interact with SRm160, associates with the oncogenic kinase Src during M phase (10, 41). The acetylated form of Sam68 has been implicated in tumor cell proliferation (1). In view of these results, it is tempting to speculate that SRm160 may play a role in influencing the malignancy of tumor cells.
It has been known that oncogenes and tumor suppressors control tumor growth and metastasis by regulating their downstream targets at the levels of transcriptional activation or repression. This study suggests that regulation of alternative splicing is yet another important mechanism in tumorigenesis. More than half of all human genes are estimated to undergo alternative splicing, some of which is regulated both temporally and spatially. The roles of CD44 isoforms in invasion clearly indicate that some of this alternative splicing is important in tumorigenesis.
Acknowledgments
We thank B. Blencowe, H. Konig, S. McCracken, and D. Shalloway for reagents and helpful suggestions; L. Aleman, J. Doench, W. Fairbrother, A. Grishok, A. Leung, A. Seila, D. Tantin, and J. Wang for discussion and critical reading of the manuscript; and J. Wang for excellent technical help.
This work was supported by U.S. Public Health Service grants R37-GM34277 (NIH) and P01-CA42063 (NCI) to P.A.S. and partially by Cancer Center Support (core) grant P30-CA14051 (NCI). C.C. is partially supported by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation.
REFERENCES
- 1.Babic, I., A. Jakymiw, and D. J. Fujita. 2004. The RNA binding protein Sam68 is acetylated in tumor cell lines, and its acetylation correlates with enhanced RNA binding activity. Oncogene 23:3781-3789. [DOI] [PubMed] [Google Scholar]
- 2.Bennett, K. L., D. G. Jackson, J. C. Simon, E. Tanczos, R. Peach, B. Modrell, I. Stamenkovic, G. Plowman, and A. Aruffo. 1995. CD44 isoforms containing exon V3 are responsible for the presentation of heparin-binding growth factor. J. Cell Biol. 128:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 4.Blencowe, B. J., R. Issner, J. A. Nickerson, and P. A. Sharp. 1998. A coactivator of pre-mRNA splicing. Genes Dev. 12:996-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, S. H., K. Takahashi, H. Eto, S. S. Yoon, and K. K. Tanabe. 2000. CD44s expression in human colon carcinomas influences growth of liver metastases. Int. J. Cancer 85:523-526. [DOI] [PubMed] [Google Scholar]
- 6.Coulter, L. R., M. A. Landree, and T. A. Cooper. 1997. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 17:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crozat, A., P. Aman, N. Mandahl, and D. Ron. 1993. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363:640-644. [DOI] [PubMed] [Google Scholar]
- 8.Eldridge, A. G., Y. Li, P. A. Sharp, and B. J. Blencowe. 1999. The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc. Natl. Acad. Sci. USA 96:6125-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fairbrother, W. G., R. F. Yeh, P. A. Sharp, and C. B. Burge. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297:1007-1013. [DOI] [PubMed] [Google Scholar]
- 10.Fumagalli, S., N. F. Totty, J. J. Hsuan, and S. A. Courtneidge. 1994. A target for Src in mitosis. Nature 368:871-874. [DOI] [PubMed] [Google Scholar]
- 11.Gunthert, U., M. Hofmann, W. Rudy, S. Reber, M. Zoller, I. Haussmann, S. Matzku, A. Wenzel, H. Ponta, and P. Herrlich. 1991. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell 65:13-24. [DOI] [PubMed] [Google Scholar]
- 12.Harn, H. J., L. I. Ho, R. Y. Shyu, J. S. Yuan, F. G. Lin, T. H. Young, C. A. Liu, H. S. Tang, and W. H. Lee. 1996. Soluble CD44 isoforms in serum as potential markers of metastatic gastric carcinoma. J. Clin. Gastroenterol. 22:107-110. [DOI] [PubMed] [Google Scholar]
- 13.Harn, H. J., L. I. Ho, C. P. Yu, M. W. Wang, H. S. Lee, J. J. Lin, W. H. Lee, N. R. Isola, and D. L. Cooper. 1994. The variant mRNA isoform of human metastasis gene (CD44V) detected in the cell lines of human hepatocellular carcinoma. Biochem. Mol. Biol. Int. 32:233-238. [PubMed] [Google Scholar]
- 14.Hofmann, M., W. Rudy, U. Gunthert, S. G. Zimmer, V. Zawadzki, M. Zoller, R. B. Lichtner, P. Herrlich, and H. Ponta. 1993. A link between ras and metastatic behavior of tumor cells: ras induces CD44 promoter activity and leads to low-level expression of metastasis-specific variants of CD44 in CREF cells. Cancer Res. 53:1516-1521. [PubMed] [Google Scholar]
- 15.Ichikawa, H., K. Shimizu, Y. Hayashi, and M. Ohki. 1994. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 54:2865-2868. [PubMed] [Google Scholar]
- 16.Jothy, S. 2003. CD44 and its partners in metastasis. Clin. Exp. Metastasis 20:195-201. [DOI] [PubMed] [Google Scholar]
- 17.Katagiri, Y. U., J. Sleeman, H. Fujii, P. Herrlich, H. Hotta, K. Tanaka, S. Chikuma, H. Yagita, K. Okumura, M. Murakami, I. Saiki, A. F. Chambers, and T. Uede. 1999. CD44 variants but not CD44s cooperate with beta1-containing integrins to permit cells to bind to osteopontin independently of arginine-glycine-aspartic acid, thereby stimulating cell motility and chemotaxis. Cancer Res. 59:219-226. [PubMed] [Google Scholar]
- 18.Konig, H., J. Moll, H. Ponta, and P. Herrlich. 1996. Trans-acting factors regulate the expression of CD44 splice variants. EMBO J. 15:4030-4039. [PMC free article] [PubMed] [Google Scholar]
- 19.Konig, H., H. Ponta, and P. Herrlich. 1998. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 17:2904-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, S. C., H. J. Harn, T. S. Lin, K. T. Yeh, Y. C. Liu, C. S. Tsai, and Y. L. Cheng. 2003. Prognostic significance of CD44v5 expression in human thymic epithelial neoplasms. Ann. Thorac. Surg. 76:213-218. [DOI] [PubMed] [Google Scholar]
- 21.Lin, Q., S. J. Taylor, and D. Shalloway. 1997. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J. Biol. Chem. 272:27274-27280. [DOI] [PubMed] [Google Scholar]
- 22.Martin, T. A., G. Harrison, R. E. Mansel, and W. G. Jiang. 2003. The role of the CD44/ezrin complex in cancer metastasis. Crit. Rev. Oncol. Hematol. 46:165-186. [DOI] [PubMed] [Google Scholar]
- 23.Matter, N., P. Herrlich, and H. Konig. 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420:691-695. [DOI] [PubMed] [Google Scholar]
- 24.McCracken, S., M. Lambermon, and B. J. Blencowe. 2002. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22:148-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCracken, S., D. Longman, I. L. Johnstone, J. F. Caceres, and B. J. Blencowe. 2003. An evolutionarily conserved role for SRm160 in 3′-end formation that functions independently of exon junction complex formation. J. Biol. Chem. 278:44153-44160. [DOI] [PubMed] [Google Scholar]
- 26.McGarvey, T., E. Rosonina, S. McCracken, Q. Li, R. Arnaout, E. Mientjes, J. A. Nickerson, D. Awrey, J. Greenblatt, G. Grosveld, and B. J. Blencowe. 2000. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J. Cell Biol. 150:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLaren, M., K. Asai, and A. Cochrane. 2004. A novel function for Sam68: enhancement of HIV-1 RNA 3′ end processing. RNA 10:1119-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meissner, M., S. Lopato, J. Gotzmann, G. Sauermann, and A. Barta. 2003. Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Exp. Cell Res. 283:184-195. [DOI] [PubMed] [Google Scholar]
- 29.Morrison, H., L. S. Sherman, J. Legg, F. Banine, C. Isacke, C. A. Haipek, D. H. Gutmann, H. Ponta, and P. Herrlich. 2001. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 15:968-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulder, J. W., P. M. Kruyt, M. Sewnath, C. A. Seldenrijk, W. F. Weidema, S. T. Pals, and G. J. Offerhaus. 1995. Difference in expression of CD44 splice variants between proximal and distal adenocarcinoma of the large bowel. Br. J. Surg. 82:1468-1470. [DOI] [PubMed] [Google Scholar]
- 31.Muller, W., A. Schneiders, K. H. Heider, S. Meier, G. Hommel, and H. E. Gabbert. 1997. Expression and prognostic value of the CD44 splicing variants v5 and v6 in gastric cancer. J. Pathol. 183:222-227. [DOI] [PubMed] [Google Scholar]
- 32.Naor, D., S. Nedvetzki, I. Golan, L. Melnik, and Y. Faitelson. 2002. CD44 in cancer. Crit. Rev. Clin. Lab. Sci. 39:527-579. [DOI] [PubMed] [Google Scholar]
- 33.Orian-Rousseau, V., L. Chen, J. P. Sleeman, P. Herrlich, and H. Ponta. 2002. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 16:3074-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira, P. A., U. Rubenthiran, M. Kaneko, S. Jothy, and A. J. Smith. 2001. CD44s expression mitigates the phenotype of human colorectal cancer hepatic metastases. Anticancer Res. 21:2713-2717. [PubMed] [Google Scholar]
- 35.Ponta, H., L. Sherman, and P. A. Herrlich. 2003. CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 4:33-45. [DOI] [PubMed] [Google Scholar]
- 36.Sherman, L., J. Sleeman, P. Dall, A. Hekele, J. Moll, H. Ponta, and P. Herrlich. 1996. The CD44 proteins in embryonic development and in cancer. Curr. Top. Microbiol. Immunol. 213:249-269. [DOI] [PubMed] [Google Scholar]
- 37.Sherman, L., D. Wainwright, H. Ponta, and P. Herrlich. 1998. A splice variant of CD44 expressed in the apical ectodermal ridge presents fibroblast growth factors to limb mesenchyme and is required for limb outgrowth. Genes Dev. 12:1058-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sleeman, J. P., K. Kondo, J. Moll, H. Ponta, and P. Herrlich. 1997. Variant exons v6 and v7 together expand the repertoire of glycosaminoglycans bound by CD44. J. Biol. Chem. 272:31837-31844. [DOI] [PubMed] [Google Scholar]
- 39.Sleeman, J. P., U. Rahmsdorf, A. Steffen, H. Ponta, and P. Herrlich. 1998. CD44 variant exon v5 encodes a tyrosine that is sulphated. Eur. J. Biochem. 255:74-80. [DOI] [PubMed] [Google Scholar]
- 40.Tacke, R., M. Tohyama, S. Ogawa, and J. L. Manley. 1998. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell 93:139-148. [DOI] [PubMed] [Google Scholar]
- 41.Taylor, S. J., and D. Shalloway. 1994. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature 368:867-871. [DOI] [PubMed] [Google Scholar]
- 42.Um, S. J., S. Y. Lee, E. J. Kim, J. Myoung, S. E. Namkoong, and J. S. Park. 2002. Down-regulation of human papillomavirus E6/E7 oncogene by arsenic trioxide in cervical carcinoma cells. Cancer Lett. 181:11-22. [DOI] [PubMed] [Google Scholar]
- 43.van der Voort, R., T. E. Taher, V. J. Wielenga, M. Spaargaren, R. Prevo, L. Smit, G. David, G. Hartmann, E. Gherardi, and S. T. Pals. 1999. Heparan sulfate-modified CD44 promotes hepatocyte growth factor/scatter factor-induced signal transduction through the receptor tyrosine kinase c-Met. J. Biol. Chem. 274:6499-6506. [DOI] [PubMed] [Google Scholar]
- 44.von Lindern, M., M. Fornerod, S. van Baal, M. Jaegle, T. de Wit, A. Buijs, and G. Grosveld. 1992. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol. Cell. Biol. 12:1687-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weg-Remers, S., H. Ponta, P. Herrlich, and H. Konig. 2001. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 20:4194-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, S. T., G. H. Sun, D. S. Hsieh, A. Chen, H. I. Chen, S. Y. Chang, and D. Yu. 2003. Correlation of CD44v5 expression with invasiveness and prognosis in renal cell carcinoma. J. Formos. Med. Assoc. 102:229-233. [PubMed] [Google Scholar]
- 47.Zhu, J., J. Shendure, R. D. Mitra, and G. M. Church. 2003. Single molecule profiling of alternative pre-mRNA splicing. Science 301:836-838. [DOI] [PubMed] [Google Scholar]