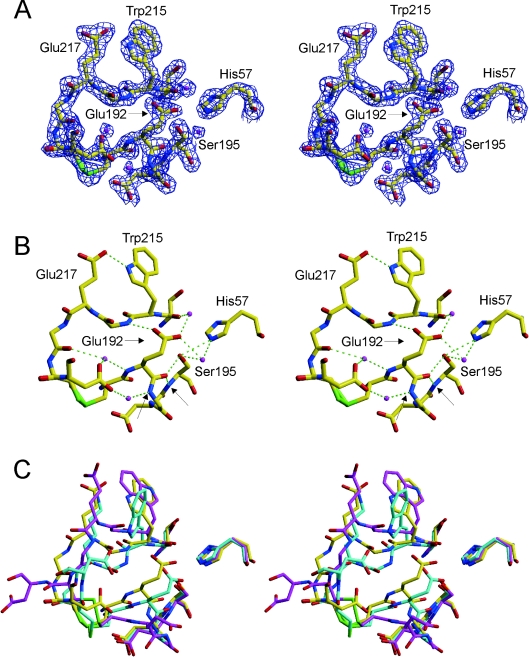
Figure 2. The active site of Na+-free wild-type thrombin is non-catalytic.
(A) The stereo representation of electron density (blue) surrounding residues 57, 191–195 and 214–221 contoured at 2σ indicates the high quality of the structure in the active-site region. Carbon atoms are shown in yellow, oxygen in red, nitrogen in blue, sulphur in green, and waters are magenta. (B) The hydrogen-bonding interactions in the active site are extensive and non-catalytic (green broken lines). Of particular importance for the activity of thrombin is the movement of Glu192, which results in a main-chain hydrogen bond to the side-chain Hγ and main-chain N–H of Ser195, and the flipping of the oxyanion hole partner Gly193. The atoms which normally constitute the oxyanion hole (Gly193 and Ser195 nitrogens) are indicated by arrows. (C) A superposition of the same active-site residues for Na+-free wild-type thrombin (yellow), recombinant slow thrombin E217K variant (cyan) and normal fast thrombin (magenta) illustrates the radical nature of the active site perturbations caused by the absence of Na+. The active-site geometry is similarly perturbed for the E217K variant, but the perpendicular orientation of the side chain of Trp215 further blocks the active site. The orientation of the Trp215 side chain in Na+-free thrombin is parallel to that of fast thrombin, but has flipped 180° to form a hydrogen bond with Glu217 (see B).