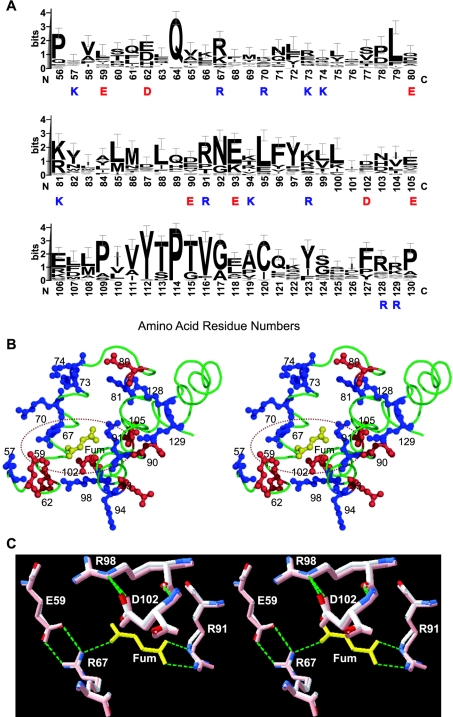
Figure 5. Fumarate binding region of human m-NAD-MDH.
(A) Sequence alignments around the fumarate binding site of the 35 decarboxylating MDHs with amino acid sequences available. Amino acid sequences of the enzymes were searched by Blast [30] and alignments were generated by Clustal W [31]. The results are expressed by sequence logos with error bars [32]. The amino acid residues highlighted are charged residues in the fumarate binding region; blue for positively and red for negatively charged amino acids. (B) Local structural representation of the charged amino acid residues at the fumarate (Fum) binding region. Blue indicates basic residues (Lys and Arg) and red indicates acidic residues (Asp and Glu). This figure was generated with PyMOL (DeLano Scientific LLC). (C) Superimposition of the closed form I (PDB code 1do8; in pink) and closed form II (PDB code 1pj4) structures, indicating the location and conformation of Asp102 in the fumarate binding pocket. The green dashed lines representing hydrogen bonds between amino acid residues and fumarate (in yellow) were generated by Swiss-Pdb Viewer [33].