Abstract
Penicillin-binding proteins (PBPs), which are the lethal targets of β-lactam antibiotics, catalyse the final stages of peptidoglycan biosynthesis of the bacterial cell wall. PBP 5 of Escherichia coli is a D-alanine CPase (carboxypeptidase) that has served as a useful model to elucidate the catalytic mechanism of low-molecular-mass PBPs. Previous studies have shown that modification of Cys115 with a variety of reagents results in a loss of CPase activity and a large decrease in the rate of deacylation of the penicilloyl–PBP 5 complex [Tamura, Imae and Strominger (1976) J. Biol. Chem. 251, 414–423; Curtis and Strominger (1978) J. Biol. Chem. 253, 2584–2588]. The crystal structure of wild-type PBP 5 in which Cys115 fortuitously had formed a covalent adduct with 2-mercaptoethanol was solved at 2.0 Å (0.2 nm) resolution, and these results provide a structural rationale for how thiol-directed reagents lower the rate of deacylation. When compared with the structure of the unmodified wild-type enzyme, a major change in the architecture of the active site is observed. The two largest differences are the disordering of a loop comprising residues 74–90 and a shift in residues 106–111, which results in the displacement of Ser110 of the SXN active-site motif. These results support the developing hypothesis that the SXN motif of PBP 5, and especially Ser110, is intimately involved in the catalytic mechanism of deacylation.
Keywords: catalytic mechanism, D-alanine carboxypeptidase, deacylation, Escherichia coli, penicillin-binding protein, X-ray crystallography
Abbreviations: 2ME, 2-mercaptoethanol; CPase, carboxypeptidase; HMM, high molecular mass; LMM, low molecular mass; p-CMB, p-chloromercuribenzoate; PEG, poly(ethylene glycol); PBP, penicillin-binding protein; R.M.S.D., root mean square deviation
INTRODUCTION
During the final stages of cell-wall synthesis in bacteria, enzymes known as penicillin-binding proteins (PBPs) catalyse the cross-linking of peptide chains from adjacent glycan strands of nascent peptidoglycan (reviewed in [1–4]). Cross-linking renders the peptidoglycan insoluble and promotes rigidity that helps to create a barrier against increases in osmotic pressure. As their name implies, these enzymes are the lethal targets of penicillin and other β-lactam antibiotics. Penicillin exerts its antimicrobial effect by forming a long-lived covalent complex with PBPs, which prevents cross-linking of the peptide chains and leads to cell death. PBPs can be grouped into two classes: the HMM (high molecular mass) PBPs, which are considered essential for cell viability, and the LMM (low molecular mass) PBPs, which are non-essential for cell viability. Some HMM PBPs are bifunctional enzymes that catalyse both the transpeptidation of the peptide chains and the transglycosylation of the glycan strands. In contrast, most LMM PBPs are D-Ala-D-Ala CPases (carboxypeptidases), which are thought to regulate the degree of cross-linking by hydrolysing the ultimate D-alanine residue of the pentapeptide [5]. Some LMM PBPs can also catalyse transpeptidation in vitro [6,7] but it remains unclear as to whether this activity also occurs in vivo.
All PBPs are characterized by a set of three conserved sequence motifs: the SXXK tetrad, which contains the active-site serine residue that forms a covalent bond with penicillin, the SXN triad and the KTG triad [8]. These motifs are widely separated in the primary structure, but come together in the folded protein to form the active site. Superimpositions of the active sites of the PBPs of known structure reveal that the active site of one PBP looks very much like another. Accordingly, the mechanism of catalysis is likely to be the same within the family of PBPs. PBPs catalyse a two-step reaction comprising acylation and deacylation. In the acylation step, the enzyme provides a general base that pulls a proton from the hydroxy group of a conserved serine residue (in the SXXK motif), facilitating a nucleophilic attack of the serine residue on the carbonyl carbon of the pentapeptide substrate. This attack results in the release of the ultimate D-alanine residue and formation of an acyl-enzyme complex between the PBP and the penultimate D-alanine residue of the peptide chain. The deacylation reaction in PBPs differs depending on whether the PBP is a transpeptidase (the HMM PBPs) or a CPase (the LMM PBPs). In transpeptidases, deacylation involves nucleophilic attack by a free amino group of the acceptor peptide side chain on to the carbonyl carbon of the acyl-enzyme complex, whereas in CPases a water molecule acts as the acceptor and the result is a tetrapeptide product.
PBP 5 from Escherichia coli is one of the best-studied PBPs and is proving to be an excellent model for probing the catalytic mechanism of PBPs. PBP 5 is a D-alanine CPase and, although it is not an essential enzyme, it is important for maintaining correct bacterial cell morphology [9,10]. Biochemical and structural studies of both wild-type and mutant forms of this enzyme have shed light on its catalytic mechanism [11,12]. For the acylation step, we have proposed that Lys47 of the SXXK tetrad functions as the general base that abstracts a protein from the reactive serine, Ser44 [13]. In contrast, a comprehensive understanding of deacylation has proven more difficult. Indeed, a commonly accepted mechanism of deacylation for all PBPs remains elusive (see, e.g., [14,15]).
In order to probe the mechanism of deacylation in PBP 5, two independent methods, each of which generates an enzyme defective in deacylation, have proven useful. The first of these is a mutant of PBP 5 (termed PBP 5′) deficient in both DD-CPase activity and deacylation of the penicilloyl–PBP complex [16]. The 30-fold increase in the half-life of the acyl-enzyme complex with penicillin G [17] is due to a G105D point mutation [18]. Comparison of the crystal structures of both PBP 5′ [11] and the wild-type enzyme [12] showed that the major difference between the two enzymes is the disordering of residues 74–90 in the mutant enzyme, which forms a loop that is adjacent to the active site. The loss of an interaction between serine residues 86 and 87 on this loop and the SXN motif in the mutant enzyme pointed to a critical role for the SXN motif in deacylation [12].
The second approach to create a deacylation-defective variant of PBP 5 is to treat the wild-type enzyme with thiol-directed reagents. Much like PBP 5′, thiol derivatization appears to affect enzyme activity non-symmetrically by inhibiting deacylation, while acylation is less affected [19,20]. PBP 5 contains only one cysteine (Cys115) and it was proposed that this residue might be involved in hydrolysis of the acyl-enzyme complex [21,22]. However, mutating this residue to serine and alanine led to no significant differences in the rate of penicilloyl-enzyme hydrolysis or in CPase activity [23]. Thus an alternative hypothesis was proposed in which modification of Cys115 with bulky thiol reagents might perturb the three-dimensional structure of the enzyme, leading to alterations in the architecture of the active site [23]. In the absence of any structural data from PBP 5 in which Cys115 has been alkylated, however, this hypothesis could not be examined.
In this study, we present the crystal structure of wild-type PBP 5 in which Cys115 has been covalently modified by 2ME (2-mercaptoethanol). When compared with the wild-type enzyme, a large conformational change in the SXN motif is observed as well as disordering of the 74–90 loop. These results suggest that both the G105D mutation and cysteine modification generate a deacylation-defective phenotype in PBP 5 ultimately by affecting the same region of the active site, namely the SXN motif.
EXPERIMENTAL
Crystallization
A soluble construct of wild-type PBP 5 (sPBP 5) was expressed and purified as described previously [23]. The protein was stored at a concentration of 8.4 mg/ml in 20 mM Tris/HCl (pH 7.5), 0.15 M NaCl and 10 mM 2ME. Crystals of sPBP 5 did not grow using the same solution conditions as for the G105D mutant (PBP 5′) [17]; hence a new crystallization search was undertaken. For this, a wide variety of solutions were tested, including Crystal Screens I and II from Hampton Research (Aliso Viejo, CA, U.S.A.). Some co-crystallization experiments with various β-lactam antibiotics were also attempted. All of the trials were conducted using the hanging-drop vapour diffusion method in which 4 μl of protein was mixed with 4 μl of well solution on a coverslip and suspended over a 1 ml reservoir. Dishes were stored at either 18 or 21 °C.
Data collection
Crystals of PBP 5 were prepared for data collection by cryoprotecting in a solution of mother liquor {24% PEG [poly(ethylene glycol)] 8000, 50 mM sodium citrate, pH 5.5, and 100 mM magnesium acetate} and glycerol. The glycerol concentration was gradually increased from 2 to 25% glycerol over a 3 h time span. Crystals were then flash-cooled in situ at 100 K and data were collected using a Rigaku RAXIS IV++ image plate system (Rigaku-MSC) mounted on a Rigaku RU-H3R X-ray generator equipped with Osmic optics and operating at 50 kV and 100 mA. A total of 150 images were collected in 1° oscillations with 5 min exposure time per frame. The crystal-to-plate distance was 150 mm. Data were processed using CrystalClear [24].
Refinement
Since the crystals generated were, in fact, of the same cell dimensions and space group as those for PBP 5′, the refinement of the structure commenced with the co-ordinates of PBP 5′, determined previously at 2.3 Å resolution [11]. After an initial refinement performed with the CNS [25], 2(|Fobs|−|Fcalc|) and |Fobs|−|Fcalc| maps were calculated and used to make manual adjustments to the model using the O program [26]. Further rounds of refinement were carried out with REFMAC interspersed with manual revision on O. Water molecules were introduced if suitable peaks of density were visible in both maps and if they had at least two potential hydrogen-bonding partners. Two residues (Glu18 and Asn208) were modelled with two side-chain conformations. Examination of the structure revealed a molecule of 2ME covalently bound to Cys115 (see the Results section); hence this structure is herein referred to as 2ME-modified PBP 5. The stereochemistry of the final model was evaluated using PROCHECK [27] and the numbering of the amino acids corresponds to the sequence of the mature processed protein.
Diffraction data for the mercury-modified PBP 5′ were reported previously [11]. These data were collected from crystals of PBP 5′ that had been soaked in mercury(II) cyanide and are 95.6% complete to 2.5 Å resolution. This structure was refined in the same manner as for the 2ME-modified PBP 5, except that all side chains were modelled with a single position and fewer water molecules were included in the final model.
Co-ordinates and structure factors of 2ME-modified PBP 5 (1NZU) and of the mercury-modified PBP 5′ (1SDN) have been deposited in the Protein Data Bank.
RESULTS
Crystallization and structure determination of 2ME-modified PBP 5
Our initial structural studies of PBP 5 began with PBP 5′, a deacylation-deficient G105D mutant form of PBP 5, which formed bullet-shaped crystals in 18–24% PEG 6000 or 8000, 150 mM NaCl and 50 mM Tris/HCl (pH 7) [11]. Attempts to crystallize the wild-type protein under similar conditions were not successful, but ultimately they were obtained in solutions containing 8% PEG 400 buffered with 50 mM Tris/HCl (pH 8) [12]. The wild-type PBP 5 crystals were of different morphology than those of PBP 5′ and belonged to a different space group (C2 for wild-type versus P32 for PBP 5′). However, during the search for different crystallization conditions, we also obtained crystals of wild-type PBP 5 in wells containing 24% PEG 8000, 50 mM sodium citrate (pH 5.5) and 100 mM magnesium acetate. These are similar to the conditions for PBP 5′ but at a lower pH. The protein drop also contained 1 mM penicillin G, but this antibiotic was not seen in the final electron-density map (not shown). The crystals appeared after 3–4 days incubation at 22 °C and were of the same characteristic bullet morphology as crystals of PBP 5′ [17].
Diffraction studies confirmed that the new crystals were indeed isomorphous with those of PBP 5′ and belonged to space group P32 with cell dimensions a=b=50.3 Å and c=138.1 Å. The diffraction data were 99.9% complete at a resolution of 2.0 Å, and these were used to refine the structure to a final R factor of 21.9% (Rfree=26.6%) (Table 1). A Ramachandran plot indicated that 91.7% of the residues were in the most favoured regions and 8.0% were in additionally allowed regions. Parts of the molecule corresponding to regions of poor electron density were omitted from the model. These are residues 1 and 2 at the N-terminus, the last residue at the C-terminus and a surface loop comprising residues 79–84.
Table 1. Data collection and refinement statistics for the 2ME-modified wild-type PBP 5 and mercury-derivatized PBP 5′.
Numbers in parentheses are for the outer shell of data. HgCN, mercury(II) cyanide. Rmerge is defined as Rmerge=ΣhklΣi |Ii(h k l)−〈I(h k l)〉|/ΣhklΣi Ii(h k l)].
| Structure | 2ME-modified wild type | PBP 5′–HgCN |
|---|---|---|
| Data collection | ||
| Resolution range (Å) | 41.5–2.0 (2.07–2.0) | 25.0–2.5 (2.59–2.5) |
| No. of measured reflections | 74362 | 34123 |
| No. of unique reflections | 26382 | 13482 |
| Completeness (%) | 99.9 (99.8) | 95.6 (88.7) |
| Mean I/σI | 6.1 (2.0) | 11.2 (4.4) |
| Rmerge (%) | 6.5 (30.8) | 7.4 (20.3) |
| Refinement | ||
| Resolution range (Å) | 15.0–2.0 | 15.0–2.5 |
| No. of reflections | 23623 | 13044 |
| No. of protein atoms | 2681 | 2679 |
| No. of solvent molecules | 174 | 46 |
| R.M.S.D. from ideal stereochemistry | ||
| Bond lengths (Å) | 0.009 | 0.011 |
| Bond angles (°) | 1.33 | 1.27 |
| Mean B value (main chain) (Å3) | 31.8 | 29.4 |
| R.M.S.D. in main chain B factors (Å3) | 0.47 | 0.59 |
| Mean B value (side chains) (Å3) | 33.4 | 31.3 |
| R.M.S.D. in side chain B factors (Å3) | 1.26 | 1.46 |
| Mean B value waters (Å3) | 35.6 | 26.4 |
| Crystallographic R factor (%) | 21.9 | 17.4 |
| Rfree (%) | 26.6 | 24.1 |
| Ramachandran plot | ||
| Residues in most favoured region (%) | 91.7 | 90.7 |
| Residues in additionally allowed regions (%) | 8.0 | 8.3 |
| Residues in generously allowed regions (%) | 0.3 | 1.0 |
| Residues in disallowed regions (%) | 0.0 | 0.0 |
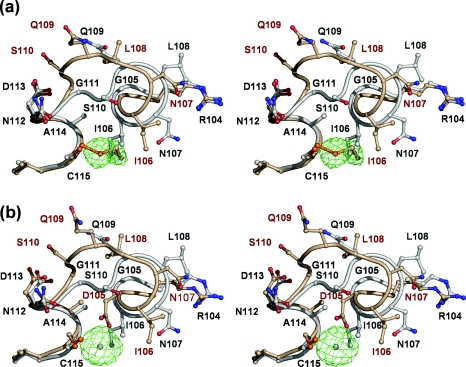
Careful scrutiny of the |Fobs|−|Fcalc| map revealed positive electron density adjacent to the thiol group of Cys115, suggesting that residue was covalently modified (Figure 1a). The size and shape of this additional density is consistent with this being a molecule of 2ME. This modification had probably occurred during the protein preparation in which 100 mM 2ME was present during elution of PBP 5 from an ampicillin affinity column [23] and the length of time that had elapsed between initial purification of the protein and the screening for crystallization conditions (∼21/2 years). Although 2ME is typically added to protein solutions to protect against oxidation of cysteine residues, there are examples of crystal structures in which it has had the opposite effect [28,29].
Figure 1. Structural effect in PBP 5 of covalent modification of Cys115 by 2ME and in PBP 5′ by mercury.
In both stereo views, residues 105–115 of wild-type PBP 5 (coloured grey) are shown superimposed on to the same residues in (a) the wild-type PBP 5 (coloured gold) in which Cys115 is modified by 2ME and (b) PBP 5-G105D (PBP 5′) (also coloured gold) in which Cys115 is modified by a mercury atom. In both cases, the same shift in residues 106–111 occurs due to the presence of 2ME or mercury at position 115 abutting Ile106. In both cases, the |Fo|−|Fc| difference electron density corresponding to 2ME or mercury is shown in green (calculated after refinement against co-ordinates in which these moieties were absent) and is contoured at 2 and 9 sigma, respectively. This Figure was prepared using PYMOL (http://pymol.sourceforge.net).
Structural description of 2ME-modified PBP 5
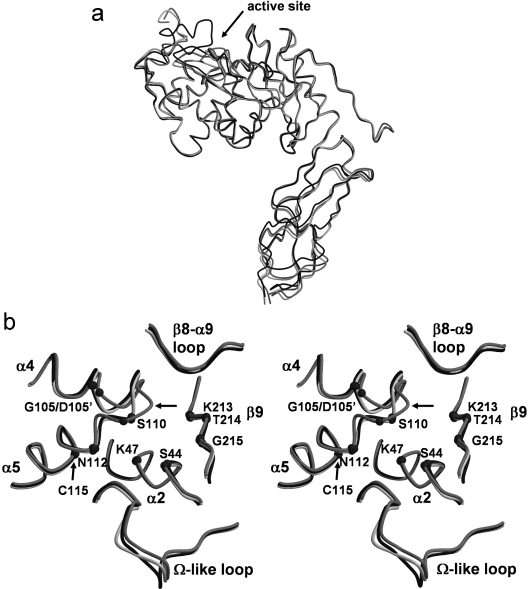
The overall fold of 2ME-modified PBP 5 is essentially the same as the wild-type [12] and PBP 5′ structures [11]. Briefly, PBP 5 comprises two domains. Domain 1 has a typical transpeptidase/penicillin-binding fold found in all PBPs and contains the active site, whereas domain 2 is mostly β-structure and may serve simply to project the active domain away from the cytoplasmic membrane and towards the peptidoglycan substrate. Superimposition of all the Cα atoms of this structure with those of the wild-type enzyme resulted in an R.M.S.D. (root mean square deviation) of 1.2 Å, indicating the close similarity between the two structures. When only the penicillin-binding domains are compared, they superimpose with an R.M.S.D. of 0.83 Å (Figure 2a). Similar values were obtained when the structures of Cys115-modified PBP 5 and PBP 5′ were compared.
Figure 2. Structural comparison of wild-type, G105D mutant (PBP 5′) and Cys115-modified PBP 5 enzymes.
(a) A superimposition of the Cα backbones of wild-type (black), PBP 5′ (light grey) and Cys115 (2ME)-modified PBP 5 (intermediate grey), showing the similarity of the overall folds of the three structures. The arrow indicates the active-site region where most of the structural differences occur. (b) Stereo representation of the same superimposition showing structural differences in the active-site region. The Cα atoms of key residues of the three active-site motifs are shown as small spheres. Note the large change in the structure of the SXN motif in Cys115-modified PBP 5 as indicated by the arrow. This Figure was prepared with MOLSCRIPT [47] and RASTER3D [48].
All of the major structural differences are located in the active-site region. First, the 2ME-modified PBP 5 structure shows disorder in the 74–90 region, located between α3 and α4, with a total absence of density for residues 79–84 and weak electron density for residues 85–90. These same residues show a similar level of disorder in the structure of PBP 5′, whereas they are ordered in the structure of the wild-type enzyme. Secondly, differences are observed in residues 155–158 of the Ω-like loop, so named with reference to its spatial overlap with the Ω loop in class A and C β-lactamases (Figure 2b). However, since this region exhibits relatively weak density, the differences observed are more likely due to the difficulty in fitting the model in this region. Finally, the most significant difference in the 2ME-modified enzyme, when compared with either the wild-type enzyme or PBP 5′, occurs in the vicinity of Cys115, as described below.
Cys115 projects into a hydrophobic core region between α2, α4 and α5 comprising residues Ile106, Met118, Ile54, Thr50 and the aliphatic part of the side chain of Lys47. Given this arrangement, the structural effect of adding a bulky group to Cys115 is, as expected, dramatic. The presence of 2ME covalently attached to Cys115 results in a clash with Ile106, causing the latter to be displaced (Figure 1a). In fact, the net effect is that the 2ME group replaces the side chain of Ile106 within the hydrophobic core region. This, in turn, leads to a shunt of five consecutive residues, beginning at Ile106 and continuing until Ser110. The consequence of this change is that α4 terminates one residue earlier (at Leu108 rather than at Gln109) in 2ME-modified PBP 5 than in the wild-type enzyme, such that the latter half of α4 is closer to a 3/10 helix in conformation rather than an α helix. The most significant result of this structural alteration is in the position of Ser110 of the conserved SXN active-site motif, which has moved a considerable distance away from the active-site cavity and now is exposed to solvent on the surface of the protein. In so doing, its Cα carbon has moved 5 Å and its side-chain hydroxy group has moved 10 Å. This movement dramatically alters the network of interactions connecting residues of the active site (see below). Surprisingly, even though the displacement in Ser110 is large, the correspondence between the two structures is restored very abruptly at Gly111.
Structure determination of PBP 5′ modified with mercury
The modification of Cys115 by 2ME in wild-type PBP 5 prompted us to re-examine diffraction data of a mercury derivative used to determine the crystal structure of PBP 5′ [11], to see whether a similar modification had occurred in this protein. The structure of mercury-modified PBP 5′ was generated by refining against these data, producing a model with a final R factor of 17.4% (Rfree=24.1%) (Table 1). As was the case with the Cys115-modified enzyme, a large portion of the surface loop (74–83) was not modelled because of poor electron density. A large peak of positive |Fobs|−|Fcalc| density was observed next to Cys115, confirming that this residue had been modified by mercury (Figure 1b). The mercury is positioned at a distance of 2.5 Å from the sulphur atom of Cys115 and 2.0 Å from the carboxylate oxygen of Asp105. This latter interaction is specific to PBP 5′ because, in the wild-type enzyme, this residue is a glycine. Most pertinently, though, the same shunt of residues 106–110 is observed in this structure compared with the 2ME-modified structure, demonstrating that alkylation at Cys115 either by mercury or by 2ME has the same structural effect on the protein. In fact, the 2ME-modified PBP 5 and mercury-modified PBP 5′ are highly similar: the R.M.S.D. between all main chain Cα atoms is 0.52 Å. Importantly, essentially the same perturbation occurs to the SXN motif (Figure 1b), and the hydrogen-bonding networks (not shown) are very similar in both active sites.
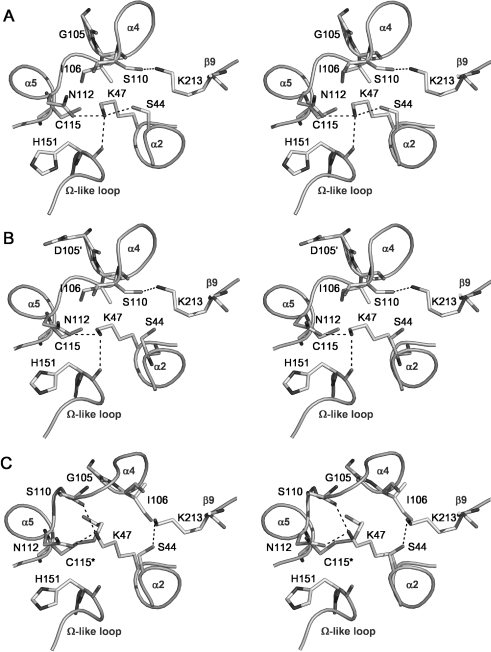
Hydrogen-bonding network in the active site
In 2ME-modified PBP 5, the movement of Ser110 has led to a major alteration in the hydrogen-bonding arrangement of the active site when compared with either wild-type PBP 5 or the PBP 5′ mutant, which have essentially the same arrangement (Figure 3). In the wild-type and PBP 5′ enzymes, the γ hydroxy group of Ser110 forms a hydrogen bond with Lys213 [11,12], but in the structure of the modified enzyme this bond is broken and Ser110 is now exposed on the surface of the molecule, beyond hydrogen-bonding distance of any other amino acid. Lys213 also loses a hydrogen bond with the carbonyl group of Ile106 (adjacent to the site of modification at Cys115), but a slight movement of its residue now permits a new interaction with Ser44.
Figure 3. Hydrogen-bonding networks in the active sites of (A) wild-type PBP 5, (B) PBP 5-G105D (PBP 5′) and (C) Cys115 (2ME)-modified PBP 5, shown as stereo representations with important residues in bond form.
Potential hydrogen bonds are shown as dashed lines. In comparison with the views of wild-type and G105D mutant PBP 5, the Cys115 modification in the 2ME-modified enzyme has caused Ser110 to be displaced away from the active site and, as a result, its hydrogen bond with Lys213 is lost and Lys213 now contacts Ser44. Note also the altered position of Lys47 in the modified enzyme, which now contacts the carbonyl of Ser110 instead of His151. This Figure was prepared using PYMOL (pymol.sourceforge.net).
The hydrogen-bonding interactions involving Lys47 are also altered. In the wild-type structure of PBP 5, the ϵ-amino group of Lys47 is within hydrogen-bonding distance of the hydroxy group of Ser44, the amide group of Asn112 and the carbonyl of His151. In the 2ME-modified enzyme, however, a slight movement of the side chains of Lys47 and Ser44 results in the breakage of the hydrogen bond between these two residues. In addition, the hydrogen bond between the ϵ-amino group of Lys47 and the carbonyl oxygen of His151 is lost and a new one has formed between the ϵ-amino group of Lys47 and the carbonyl of Ser110.
DISCUSSION
The first crystal structure for PBP 5 was of a G105D mutant, termed PBP 5′, which displays a 30-fold decrease in the rate of deacylation of the penicilloyl-enzyme complex [11,17]. When compared with the structure of the wild-type enzyme, the major difference is the disordering of residues 74–90 in PBP 5′ [12]. This disordering was attributed to the disruption of a hydrophobic core region (comprising α4, α5, β7, β8 and the 74–90 loop) by Asp105, ultimately leading to the loss of contacts between two serines (Ser86 and Ser87) present on the 74–90 loop and Ser110 and Asn112 of the SXN motif [12]. This result suggested that the primary mechanism for the defect in deacylation was the loss of order of the 74–90 region. In support of this hypothesis, deletion of the 74–90 region in PBP 5 resulted in a decrease in the rate of deacylation to the same extent as in PBP 5′ [12].
A second method to produce a deacylation-defective PBP 5 is by treatment of the enzyme with thiol-directed agents [19,20]. The likely target in PBP 5 for such agents is Cys115, which is the only cysteine residue in PBP 5. Interestingly, Cys115 lies very close to Gly105, suggesting that the defect in deacylation in both PBP 5′ and thiol-modified PBP 5 may be the result of the same mechanism. The structure of Cys115-modified PBP 5 presented here allows an examination of this hypothesis.
This new structure of PBP 5 arose from efforts to determine the crystal structure of wild-type PBP 5, culminating in a structure solved from crystals grown in space group C2 [12]. Wild-type PBP 5 crystallized under very different conditions from those for PBP 5′, which crystallized in space group P32 [17]. Surprisingly, an older preparation of the wild-type protein crystallized under similar conditions to PBP 5′, and crystals were of the same space group (P32). In this structure, Cys115 is modified by 2ME and, importantly, the 74–90 region shows a similar level of disorder as seen in PBP 5′. These results are consistent with the disordering of residues 74–90 being the primary cause of the defect in deacylation. However, such a facile interpretation is complicated by the presence of an additional structural change, the large conformational shift in the region encompassing the SXN motif, because such a dramatic change in the positions of active-site residues would be expected to have a significant impact on enzyme activity.
The SXN motif is one of the three active-site motifs that are common to all PBPs and related penicillin-interacting enzymes [3]. Following covalent modification of Cys115, the conformation of the SXN motif is altered significantly due to a large shift of Ser110 away from the active site and the loss of a hydrogen bond with Lys213 (from the KTG motif) that is present in the wild-type enzyme. Given the high degree of conservation of the SXN motif and the critical interactions made by Ser110 in the active site, it seems surprising that when PBP 5 is treated with the thiol reagent p-CMB (p-chloromercuribenzoate), which presumably is also directed against Cys115, the enzyme would show any activity at all, either acylation or deacylation. Indeed, mutation of Ser110 to Ala leads to a mutant protein with severely decreased activities for both of these reactions [30].
One explanation for this apparent discrepancy is that the shift in the SXN motif seen in the crystal structure does not occur when PBP 5 is treated with p-CMB [20]. However, since the 2ME group on Cys115 directly causes the displacement of the side chain of Ile106 from a hydrophobic core region, with the concomitant shunt of residues 106–110, this explanation seems implausible because the large size of a mercury atom would likely have the same effect. This prompted us to re-examine diffraction data collected from crystals of PBP 5′ derivatized with mercury(II) cyanide that were used to solve that structure [11]. A structure generated from refinement against these data shows that Cys115 is indeed derivatized by mercury and that the same shift of the SXN motif had occurred (Figure 1b). Unfortunately, attempts to solve the structure of wild-type PBP 5 treated with mercury derivatives (either by soaking of the crystals or by co-crystallization trials) were unsuccessful. Furthermore, the serendipitous nature of the derivatization of the wild-type enzyme with 2ME precludes reproducing this protein preparation for the purposes of determining the kinetic properties of a 2ME-modified PBP 5. Nevertheless, although a direct correlation between the crystal structure of Cys115-modified PBP 5 and kinetic data is not possible, it is reasonable to assume that the same shift in the SXN motif occurred in wild-type PBP 5 when treated with p-CMB.
How then can such an enzyme retain activity for acylation by penicillin? One explanation consistent with the data is that when Cys115 of PBP 5 in solution is modified by p-CMB or 2ME, the SXN motif has a propensity to exist in two conformations: a native conformation, as observed in wild-type PBP 5 and PBP 5′, and a non-native conformation, as observed in the structure of the 2ME-modified enzyme and the mercury derivative of PBP 5′. However, when the protein is organized within a crystal lattice, only the non-native form is trapped. When the SXN motif occupies a position resembling the native protein, the enzyme can still be acylated by penicillin while the acyl-enzyme complex remains defective for deacylation due to disorder of the 74–90 loop. Hence, by switching between the two conformations, p-CMB-treated PBP 5 retains some activity for acylation, whereas the S110A mutant is essentially inactive because it has lost a functional group that mediates important hydrogen-bonding interactions. Such a hypothesis is consistent with current ideas regarding the catalytic mechanism of PBP 5, because acylation is believed to be mediated by Lys47 [13], which moves relatively little in the Cys115-modified enzyme, whereas Ser110 is thought to play a major role in deacylation (see below).
Interestingly, a localized disordering of the equivalent region was observed in the structure of a penicillin-resistant variant of PBP 2x from Streptococcus pneumoniae that also caused a displacement of Ser395 (equivalent to Ser110 in PBP 5) away from the active site [31]. The fact that the same region is ordered in the penicillin-sensitive form of the enzyme [32,33] suggests that an alteration of structure in this region may in fact have an effect on acylation by β-lactam antibiotics. However, the PBP 2x system is complicated by the large number of mutations between the penicillin-sensitive and -resistant forms of the enzyme, making it difficult to dissect the individual contributions of each of the 83 amino acid substitutions towards the altered enzyme activity. Nevertheless, it does show that, under certain conditions, the region prior to the SXN motif can change structure in other PBPs.
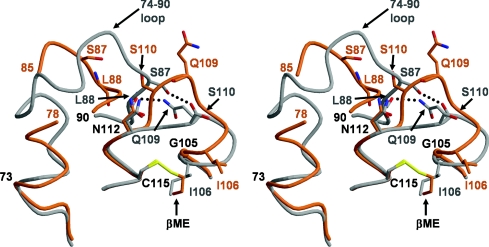
Another issue that requires explanation is how modification of Cys115 leads to disorder in the 74–90 region of the modified enzyme. One possibility is that the flexibility of this loop is simply a property of different crystal packing interactions in the P32 crystal form of PBP 5, as opposed to those in the C2 form of crystals used to determine the structure of the wild-type enzyme (in which the 74–90 loop is well ordered). Arguing against this, though, a structure of PBP 5′ solved from crystals of space group P1, which manifests a wholly different crystal packing arrangement compared with the same enzyme solved in the P32 space group, also shows the same disordering of the 74–90 loop (results not shown). Thus these results suggest that the disordering of this region exists before crystallization of the protein. In the crystal structure of PBP 5′, the mechanism of disordering was clear: the aspartate residue at position 105 (in place of glycine in the wild-type enzyme) clashed with Leu88 and Met88, displacing these residues and the surrounding 74–90 region [12]. A similar mechanism is less obvious in the Cys115-modified enzyme, because there is a glycine residue at position 105 and the presence of 2ME has not directly affected any residue other than Ile106. However, the shunting of residues 106–110 leads to the loss of hydrogen bonds between the side chain of Gln109 and the carbonyl group of Leu88 and between the carbonyl group of Gln109 and Ser87, and the loss of these interactions may be responsible for the disordering of the 74–90 loop (Figure 4). Hence, although the routes may be different, modification of Cys115 by 2ME in wild-type PBP 5 or mutation of Gly105 to Asp in PBP 5′ both lead to a disordering of the 74–90 region. Because the 2ME at Cys115 clashes with Ile106, this modification has the additional effect of altering residues 106–110 with a dramatic change in the structure of the SXN motif, whereas an aspartate residue at 105 in PBP 5′ results only in disordering of the 74–90 loop.
Figure 4. The loss of two hydrogen bonds as a result of the shift in the SXN motif contributes to disordering of the 74–90 region in Cys115-modified PBP 5.
In this stereo view, wild-type PBP 5 is represented by a grey backbone and side chains, whereas 2ME-modified PBP 5 is shown in orange. In wild-type PBP 5, there are two potential hydrogen bonds (shown as dashed lines) that link the SXN motif with the 74–90 region, which is fully ordered. In 2ME-modified PBP 5, these bonds are absent due to the shift in residues 106–111 induced by the 2ME alkylating Cys115, and their loss is hypothesized to lead to destabilization and disordering of the 74–90 loop. Residues that differ significantly in position between the two structures are coloured grey for wild-type and orange for 2ME-modified PBP 5. This Figure was prepared with MOLSCRIPT [47] and RASTER3D [48].
Taken together, the three crystal structures of PBP 5 suggest that both means of producing a deacylation-defective enzyme (by either mutation at position 105 or modification of Cys115) lead to disordering of the 74–90 region and the loss of interactions with the SXN motif. Irrespective of whether the large shift in the SXN motif observed in the Cys115-modified enzyme is also linked to the defect in deacylation, all data point to a role for the SXN motif in the mechanism of deacylation.
The catalytic mechanism underlying the enzymatic activity of PBPs remains a subject of debate. The reaction comprises two steps, acylation and deacylation, and neither of these are understood at the mechanistic level. In keeping with the mechanism first proposed for class A β-lactamases [34] and later extended to PBPs [15,35,36], one hypothesis for the mechanism of acylation in PBP 5 is that Lys47 is the general base that abstracts a proton from Ser44, activating the latter for nucleophilic attack on the carbonyl carbon of the penultimate D-Ala of the peptide substrate [13]. However, an alternative mechanism involving transfer of the proton from the serine nucleophile to the serine residue of the SXN motif, with concomitant transfer of a proton from the serine residue of the SXN motif to the β-lactam leaving group, has also been discussed in the context of other PBPs [33,37–39].
Even less is known about the mechanism of deacylation in PBPs. Initial hypotheses focused on a loop in the active site of PBPs that is spatially equivalent to the Ω loop of class A β-lactamases, because in these enzymes this loop contains both Glu166 and Asn170 that are believed to orientate a hydrolytic water molecule during deacylation [40–43]. It was suggested, first for PBP 5 [11], and later for pneumococcal PBP 2x [44] and the K15 DD-transpeptidase [15], that the equivalent loop may contain the catalytic machinery for deacylation. Subsequent structural and mutagenesis data in PBP 5, however, suggested instead that the SXN motif, and not the Ω-like loop, is intimately involved in deacylation [12,45]. Based on a crystal structure of PBP 5 in complex with the peptide-boronic acid, we have proposed a mechanism for deacylation in which Ser110 polarizes the hydrolytic water molecule through a hydrogen-bonding network comprising Ser87, Ser110, Lys213 and a bridging water molecule [45]. The present work, which points to an altered environment of the SXN motif in deacylation-defective variants of PBP 5 and, in particular, the loss of a hydrogen bond between Ser110 and Lys213, supports this hypothesis. Moreover, the equivalent motif of the SXN triad in the R61 DD-peptidase, which contains tyrosine in place of serine, is also believed to be responsible for deacylation in that enzyme [46]. In PBP 5 that is defective in deacylation, the loss of critical interactions between the SXN motif and Ser86 and Ser87 impairs the ability of Ser110 to function within this network. It appears that a similar mechanism is at play when deacylation in PBP 5 is impaired by treatment of the enzyme with alkylating agents.
Acknowledgments
This work was supported by the National Institutes of Health grants GM66861 to C.D. and AI36901 to R.A.N.
References
- 1.Waxman D. J., Strominger J. L. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu. Rev. Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 2.Georgopapadakou N. H., Smith S. A., Cimarusti C. M., Sykes R. B. Binding of monobactams to penicillin-binding proteins of Escherichia coli and Staphylococcus aureus: relation to antibacterial activity. Antimicrob. Agents Chemother. 1983;23:98–104. doi: 10.1128/aac.23.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghuysen J. M. Serine beta-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 4.Goffin C., Ghuysen J. M. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morlot C., Noirclerc-Savoye M., Zapun A., Dideberg O., Vernet T. The D,D-carboxypeptidase PBP3 organizes the division process of Streptococcus pneumoniae. Mol. Microbiol. 2004;51:1641–1648. doi: 10.1046/j.1365-2958.2003.03953.x. [DOI] [PubMed] [Google Scholar]
- 6.Grandchamps J., Nguyen-Disteche M., Damblon C., Frere J. M., Ghuysen J. M. Streptomyces K15 active-site serine DD-transpeptidase: specificity profile for peptide, thiol ester and ester carbonyl donors and pathways of the transfer reactions. Biochem. J. 1995;307:335–339. doi: 10.1042/bj3070335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamin M., Wilkin J. M., Frere J. M. A new kinetic mechanism for the concomitant hydrolysis and transfer reactions catalyzed by bacterial DD-peptidases. Biochemistry. 1993;32:7278–7285. doi: 10.1021/bi00079a026. [DOI] [PubMed] [Google Scholar]
- 8.Ghuysen J. M. Molecular structures of penicillin-binding proteins and beta-lactamases. Trends Microbiol. 1994;2:372–380. doi: 10.1016/0966-842x(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 9.Nelson D. E., Young K. D. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 2000;182:1714–1721. doi: 10.1128/jb.182.6.1714-1721.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson D. E., Young K. D. Contributions of PBP 5 and DD-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 2001;183:3055–3064. doi: 10.1128/JB.183.10.3055-3064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies C., White S. W., Nicholas R. A. Crystal structure of a deacylation-defective mutant of penicillin-binding protein 5 at 2.3 Å resolution. J. Biol. Chem. 2001;276:616–623. doi: 10.1074/jbc.M004471200. [DOI] [PubMed] [Google Scholar]
- 12.Nicholas R. A., Krings S., Tomberg J., Nicola G., Davies C. Crystal structure of wild-type penicillin-binding protein 5 from E. coli: implications for deacylation of the acyl-enzyme complex. J. Biol. Chem. 2003;278:52826–52833. doi: 10.1074/jbc.M310177200. [DOI] [PubMed] [Google Scholar]
- 13.Stefanova M. E., Davies C., Nicholas R. A., Gutheil W. G. pH, inhibitor, and substrate specificity studies on Escherichia coli penicillin-binding protein 5. Biochim. Biophys. Acta. 2002;1597:292–300. doi: 10.1016/s0167-4838(02)00311-4. [DOI] [PubMed] [Google Scholar]
- 14.McDonough M. A., Anderson J. W., Silvaggi N. R., Pratt R. F., Knox J. R., Kelly J. A. Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins. J. Mol. Biol. 2002;322:111–122. doi: 10.1016/s0022-2836(02)00742-8. [DOI] [PubMed] [Google Scholar]
- 15.Rhazi N., Charlier P., Dehareng D., Engher D., Vermeire M., Frere J. M., Nguyen-Disteche M., Fonze E. Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis. Biochemistry. 2003;42:2895–2906. doi: 10.1021/bi027256x. [DOI] [PubMed] [Google Scholar]
- 16.Matsuhashi M., Maruyama I. N., Takagaki Y., Tamaki S., Nishimura Y., Hirota Y. Isolation of a mutant of Escherichia coli lacking penicillin-sensitive D-alanine carboxypeptidase IA. Proc. Natl. Acad. Sci. U.S.A. 1978;75:2631–2635. doi: 10.1073/pnas.75.6.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholas R. A., Strominger J. L. Relations between beta-lactamases and penicillin-binding proteins: beta-lactamase activity of penicillin-binding protein 5 from Escherichia coli. Rev. Infect. Dis. 1988;10:733–738. doi: 10.1093/clinids/10.4.733. [DOI] [PubMed] [Google Scholar]
- 18.Broome-Smith J., Spratt B. G. An amino acid substitution that blocks the deacylation step in the enzyme mechanism of penicillin-binding protein 5 of Escherichia coli. FEBS Lett. 1984;165:185–189. doi: 10.1016/0014-5793(84)80166-0. [DOI] [PubMed] [Google Scholar]
- 19.Tamura T., Imae Y., Strominger J. L. Purification to homogeneity and properties of two D-alanine carboxypeptidases I from Escherichia coli. J. Biol. Chem. 1976;251:414–423. [PubMed] [Google Scholar]
- 20.Curtis S. J., Strominger J. L. Effects of sulphydryl reagents on the binding and release of penicillin G by D-alanine carboxypeptidase IA of Escherichia coli. J. Biol. Chem. 1978;253:2584–2588. [PubMed] [Google Scholar]
- 21.Amanuma H., Strominger J. L. Purification and properties of penicillin-binding proteins 5 and 6 from the dacA mutant strain of Escherichia coli (JE 11191) J. Biol. Chem. 1984;259:1294–1298. [PubMed] [Google Scholar]
- 22.Nicholas R. A., Ishino F., Park W., Matsuhashi M., Strominger J. L. Purification and sequencing of the active site tryptic peptide from penicillin-binding protein 5 from the dacA mutant strain of Escherichia coli (TMRL 1222) J. Biol. Chem. 1985;260:6394–6397. [PubMed] [Google Scholar]
- 23.Nicholas R. A., Strominger J. L. Site-directed mutants of a soluble form of penicillin-binding protein 5 from Escherichia coli and their catalytic properties. J. Biol. Chem. 1988;263:2034–2040. [PubMed] [Google Scholar]
- 24.Pflugrath J. W. The finer things in X-ray diffraction data collection. Acta Crystallogr. D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 25.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Jones T. A., Zou J.-Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein structures in electron-density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 28.Vaughn D. E., Rodriguez J., Lazebnik Y., Joshua-Tor L. Crystal structure of Apaf-1 caspase recruitment domain: an alpha-helical Greek key fold for apoptotic signaling. J. Mol. Biol. 1999;293:439–447. doi: 10.1006/jmbi.1999.3177. [DOI] [PubMed] [Google Scholar]
- 29.Chumanevich A. A., Krupenko S. A., Davies C. The crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase: mechanism of hydrolysis and its interplay with the dehydrogenase domain. J. Biol. Chem. 2004;279:14355–14364. doi: 10.1074/jbc.M313934200. [DOI] [PubMed] [Google Scholar]
- 30.van der Linden M. P., de Haan L., Dideberg O., Keck W. Site-directed mutagenesis of proposed active-site residues of penicillin-binding protein 5 from Escherichia coli. Biochem. J. 1994;303:357–362. doi: 10.1042/bj3030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dessen A., Mouz N., Gordon E., Hopkins J., Dideberg O. Crystal structure of PBP2x from a highly penicillin-resistant Streptococcus pneumoniae clinical isolate: a mosaic framework containing 83 mutations. J. Biol. Chem. 2001;276:45106–45112. doi: 10.1074/jbc.M107608200. [DOI] [PubMed] [Google Scholar]
- 32.Pares S., Mouz N., Petillot Y., Hakenbeck R., Dideberg O. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 1996;3:284–289. doi: 10.1038/nsb0396-284. [DOI] [PubMed] [Google Scholar]
- 33.Gordon E., Mouz N., Duee E., Dideberg O. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 2000;299:477–485. doi: 10.1006/jmbi.2000.3740. [DOI] [PubMed] [Google Scholar]
- 34.Strynadka N. C., Adachi H., Jensen S. E., Johns K., Sielecki A., Betzel C., Sutoh K., James M. N. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 Å resolution. Nature (London) 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 35.Lim D., Strynadka N. C. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 2002;9:870–876. doi: 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]
- 36.Thomas B., Wang Y., Stein R. L. Kinetic and mechanistic studies of penicillin-binding protein 2x from Streptococcus pneumoniae. Biochemistry. 2001;40:15811–15823. doi: 10.1021/bi011368r. [DOI] [PubMed] [Google Scholar]
- 37.Oliva M., Dideberg O., Field M. J. Understanding the acylation mechanisms of active-site penicillin-recognizing proteins: a molecular dynamics simulation study. Proteins: Struct. Funct. Genet. 2003;53:88–100. doi: 10.1002/prot.10450. [DOI] [PubMed] [Google Scholar]
- 38.Morlot C., Pernot L., Le Gouellec A., Di Guilmi A. M., Vernet T., Dideberg O., Dessen A. Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae. J. Biol. Chem. 2005;280:15984–15991. doi: 10.1074/jbc.M408446200. [DOI] [PubMed] [Google Scholar]
- 39.Sauvage E., Kerff F., Fonze E., Herman R., Schoot B., Marquette J. P., Taburet Y., Prevost D., Dumas J., Leonard G., et al. The 2.4-Å crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell. Mol. Life Sci. 2002;59:1223–1232. doi: 10.1007/s00018-002-8500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adachi H., Ohta T., Matsuzawa H. Site-directed mutants, at position 166, of RTEM-1 beta-lactamase that form a stable acyl-enzyme intermediate with penicillin. J. Biol. Chem. 1991;266:3186–3191. [PubMed] [Google Scholar]
- 41.Guillaume G., Vanhove M., Lamotte-Brasseur J., Ledent P., Jamin M., Joris B., Frere J. M. Site-directed mutagenesis of glutamate 166 in two beta-lactamases. Kinetic and molecular modeling studies. J. Biol. Chem. 1997;272:5438–5444. doi: 10.1074/jbc.272.9.5438. [DOI] [PubMed] [Google Scholar]
- 42.Delaire M., Lenfant F., Labia R., Masson J. M. Site-directed mutagenesis on TEM-1 beta-lactamase: role of Glu166 in catalysis and substrate binding. Protein Eng. 1991;4:805–810. doi: 10.1093/protein/4.7.805. [DOI] [PubMed] [Google Scholar]
- 43.Lewis E. R., Winterberg K. M., Fink A. L. A point mutation leads to altered product specificity in beta-lactamase catalysis. Proc. Natl. Acad. Sci. U.S.A. 1997;94:443–447. doi: 10.1073/pnas.94.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chesnel L., Zapun A., Mouz N., Dideberg O., Vernet T. Increase of the deacylation rate of PBP2x from Streptococcus pneumoniae by single point mutations mimicking the class A beta-lactamases. Eur. J. Biochem. 2002;269:1678–1683. doi: 10.1046/j.1432-1327.2002.02815.x. [DOI] [PubMed] [Google Scholar]
- 45.Nicola G., Peddi S., Stefanova M. E., Nicholas R. A., Gutheil W. G., Davies C. Crystal structure of Escherichia coli penicillin-binding protein 5 bound to a tripeptide boronic acid inhibitor: a role for Ser-110 in deacylation. Biochemistry. 2005;44:8207–8217. doi: 10.1021/bi0473004. [DOI] [PubMed] [Google Scholar]
- 46.Silvaggi N. R., Anderson J. W., Brinsmade S. R., Pratt R. F., Kelly J. A. The crystal structure of phosphonate-inhibited d-Ala-d-Ala peptidase reveals an analogue of a tetrahedral transition state. Biochemistry. 2003;42:1199–1208. doi: 10.1021/bi0268955. [DOI] [PubMed] [Google Scholar]
- 47.Kraulis P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 48.Merritt E. A., Murphy M. E. P. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]