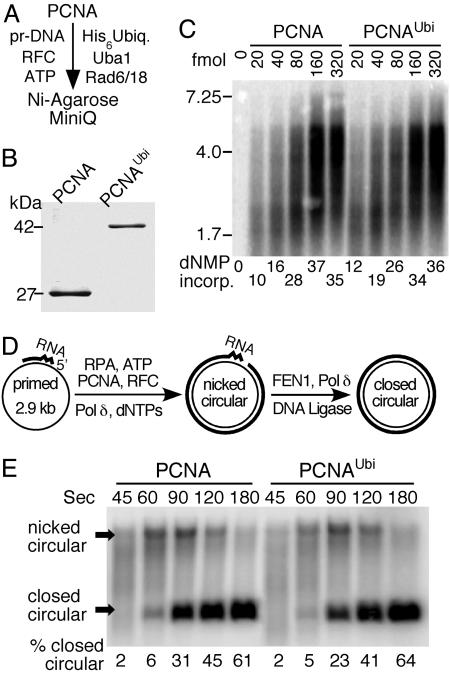
Fig. 2.
Biochemical properties of PCNAUbi in DNA replication. (A) Purification scheme. Pr-DNA is DECAprimed Bluescript SKII ssDNA. (B) Gel analysis (11% SDS/PAGE) of PCNA and purified PCNAUbi. Staining was with colloidal Coomassie blue. Scanning of the gel showed that ≈3% nonubiquitinated PCNA remained in the purified preparation. (C) Replication of RPA-coated singly primed mp18 ssDNA (60-fmol circles) by Pol δ (120 fmol) and RFC (120 fmol) with increasing PCNA or PCNAUbi was performed essentially as described (38). [α-32P]dTTP was used as radioactive tracer. The PCNA was loaded by RFC for 1 min at 30°C, Pol δ was added, and incubation continued for an additional 90 sec. Products were analyzed on a 1% alkaline agarose gel. (D) Scheme for measuring Okazaki fragment maturation kinetics. (E) Replication and maturation assays of RPA-coated circular ssDNA (100 fmol), primed with an RNA8DNA22 primer, by RFC (200 fmol), PCNA or PCNAUbi (300 fmol), Pol δ (200 fmol), FEN1 (200 fmol), and DNA ligase I (500 fmol) were exactly as described (28). Replication products after the indicated times were separated on a 1% agarose gel in the presence of 0.5 μg/ml ethidium bromide. This method separates covalently closed circular DNA from nicked circular DNA. [α-32P]dTTP was used as radioactive tracer.