Abstract
Gene targeting often results in knockout mice that show several phenotypes, some of which may not directly relate to the intrinsic function of the disrupted gene. Hence, to study the biological function of genes using knockout mice, one must identify the defects that are directly due to the loss of the targeted gene. Runx3 is a transcription factor that regulates lineage-specific gene expression in developmental processes. Recently, two groups produced Runx3 knockout mice. Two comparable defects were identified in both knockout strains, one involved neurogenesis and the other thymopoiesis. In addition, a stomach defect pertaining to gastric cancer was observed in one of the mutant strains, but not in the other. Here, we assess the differences between the two Runx3 mutant strains and discuss further studies that could reconcile these discrepancies. This article highlights the difficulties of inferring gene function through the interpretation of knockout phenotypes.
Introduction
Many knockout (KO) mice have multiple, and sometimes complex, phenotypic defects. Often, the null phenotype does not seem to recapitulate the known cellular function of the gene. This is frequently the case when the gene being studied has a distinct tissue- and/or temporal-specific function that is difficult to replicate in cell culture. Many homozygous KO mice die in utero, and those that are viable often have nutritional or immunological deficiencies that cause secondary phenotypes associated with aberrant growth and survival. Thus, the challenging aspect of analysis of gene function in KO mice is to identify the defects that are directly linked to the loss of function of the targeted gene. This study of Runx3 involvement in gastric cancer is a good example of such a challenge.
The RUNX transcription factors
Mammalian RUNX3 belongs to the runt-domain family of transcription factors that act as regulators of gene expression in several important developmental pathways. The three RUNX genes, RUNX1, RUNX2 and RUNX3, appeared early in evolution and have maintained extensive structural similarities (Fig. 1A; Eggers et al., 2002; Kalev-Zylinska et al., 2002; Levanon et al., 2003). Members of the RUNX family show homology in a 128-amino-acid region known as the runt domain (RD), which directs the binding of RUNX proteins to DNA and mediates their interaction with the protein core-binding factor-β (CBF-β; Fig. 1B) (Ito & Bae, 1997; Speck, 2001). CBF-β enhances the binding of the RUNX proteins to their target DNA and is essential for their proper function (Adya et al., 2000).
Figure 1.
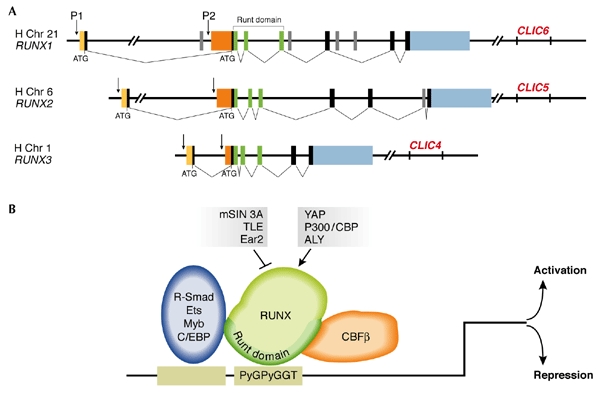
The mammalian RUNX genes: structure and mode of function. (A) The three mammalian RUNX genes have similar genomic organization with two promoters (P1 and P2) and a very large first intron. The two promoters give rise to two biologically distinct 5′ untranslated regions (UTRs) (yellow and orange). In humans and mice, each gene resides on different chromosomes (human 21, 6 and 1, and mouse 16, 17 and 4, respectively). The highly conserved runt domain is encoded by the three exons marked in green. Exons comprising the transactivation domain are shown in black and grey and the 3′ UTR in blue. Runx3 is the smallest and simplest of the three genes. (B) The runt domain directs binding to the RUNX DNA-motif PyGPyGGT at the promoter of target genes, and protein–protein interactions withcore-binding factor-β (CBF-β). The RUNX proteins bind to the same DNA motif and either activate or repress transcription through interactions with other transcription factors (blue ellipse) and co-activators (arrows), or co-repressors (blocked line). Of note, due to lack of space, only a few examples of RUNX transcriptional co-modulators are indicated.
The RUNX genes are regulated at the transcriptional level by two promoters and at the translational level by an internal ribosome-entry site (IRES) and cap-dependent translation control (Fig. 1; Miyoshi et al., 1995; Ghozi et al., 1996; Geoffroy et al., 1998; Pozner et al., 2000; Bangsow et al., 2001; Levanon et al., 2001b; Rini & Calabi, 2001; Xiao et al., 2001; Stewart et al., 2002). All RUNX proteins bind to the same DNA motif and activate or repress the transcription of their target genes through the recruitment of common transcriptional modulators (Fig. 1B; Karsenty, 2000; Wheeler et al., 2000). In spite of this, the RUNX genes have well-defined biological functions, which are reflected in their unique expression patterns (Simeone et al., 1995; North et al., 1999; Levanon et al., 2001a; Chen et al., 2002; Stricker et al., 2002; Yamashiro et al., 2002) and the distinct phenotypes that are shown by the corresponding KO mice (Otto et al., 1997; Karsenty, 2000; Speck, 2001; Inoue et al., 2002; Levanon et al., 2002; Li et al., 2002). RUNX3 is located on human chromosome 1p36.1 (Levanon et al., 1994) and seems to be the most ancient of the three RUNX genes (Bangsow et al., 2001; Levanon et al., 2003). This is consistent with its function in the neurogenesis of the monosynaptic reflex arc (Inoue et al., 2002; Levanon et al., 2002), the simplest neuronal response circuit found in the most primitive animals, such as hydra-like organisms.
Runx3, gastric mucosa hypertrophy and stomach cancer
Stomach cancer is the second most common form of malignancy and is a major contributor to cancer mortality throughout the world (Fuchs & Mayer, 1995; Parkin et al., 1999). Gastric carcinomas have been linked with the loss of homozygosity at various chromosomal loci, but no single gene that accounts for the majority of cases has been identified. Recently, Li and colleagues (Li et al., 2002) reported that the gastric mucosa of Runx3 null mice showed hyperplasia and concluded that lack of RUNX3 is causally related to human gastric cancer. Consistent with this, an analysis of RUNX3 in human stomach cancer cell lines and primary human tumours revealed hemizygosity in 40% of the tumours analysed, and silencing by promoter hypermethylation in 60% of the tumours, the latter figure rising to 90% in the advanced stage tumours (Li et al., 2002). Furthermore, silencing by hypermethylation of the Runx3 promoter was also observed in cell lines derived from N-methyl-M-nitrosourea-induced mouse stomach carcinomas (Guo et al., 2002).
These observations prompt the question: does the loss of Runx3 in the KO mice cause gastric tumorigenesis? In the case of the Runx3−/− mice described by Li et al. (2002) (hereafter referred to as Runx3 type I KO), which were inbred on a C57BL/6 background, most progeny died of starvation soon after birth and none survived for more than 10 days. Therefore, studies of tumorigenesis in these mice were not feasible. However, another strain of Runx3 KO mice (Runx3 type II KO), which were bred on a heterogeneous genetic background (ICR and MF1), produced progeny of which a significant number survived for several months (Levanon et al., 2002). Intriguingly, newborn Runx3 type II KO mice did not show hyperplasia of the gastric epithelium and did not develop gastric tumours (Levanon et al., 2002). This observation strongly suggests that loss of Runx3 is not necessarily associated with gastric neoplasia, and that the mucosal hypertrophy observed in the newborn Runx3 type I KO mice might be related to the strain on which the study was performed. It is interesting to note that the C57BL/6 strain is more susceptible to Helicobacter felis infection, which often results in a severe gastric phenotype that is characterized by increased proliferation of the mucosal epithelium (Wang et al., 1998), than other strains such as BALB/c or C3H/HeJ. Therefore, as suggested by Lund van Lohuizen (Lund van Lohuizen, 2002), the possible involvement of Helicobacter in the aetiology of stomach lesions in Runx3 KO mice should not be overlooked.
As mentioned earlier, CBF-β is essential for the activity of all the Runx proteins (Adya et al., 2000); consequently, CBF-β -deficient mice recapitulate the Runx1 null phenotype, as they lack definitive haematopoiesis and die due to haemorrhaging at embryonic day 12.5 (Adya et al., 2000; Speck, 2001). Recently, three groups produced new strains of CBF-β-mutant mice in which the early haematopoietic defect was reversed (Kundu et al., 2002; Miller et al., 2002; Yoshida et al., 2002). The progeny of these mice were born, but died within one day of birth. As the stomach defect in Runx3 type I KO mice was detected immediately after birth and before suckling commenced, it would be interesting to look for mucosal hyperplasia in these new CBF-β−/− rescue mutants. Moreover, the early death of the Runx3 KO C57BL/6 pups precluded the analysis of tumour development in these mice. However, as breeding them into an ICR background significantly extended their lifespan (Inoue et al., 2002), it is now possibile to examine stomach tumorigenesis in these mice. It would be equally interesting to breed the Runx3 type II KO mice onto a C57BL/6 background and to examine the newborns for gastric mucosa hyperplasia.
Is Runx3 intrinsically required in the gastric epithelium?
The phenotypic differences between the two Runx3 KO strains also extend to the expression pattern of Runx3 during development. Analysis of heterozygous Runx3LacZ/+ embryos of both type I and type II KO mice revealed X-gal staining in sensory ganglia, epidermal appendages and developing skeletal elements (Levanon et al., 2001a, 2002; Li et al., 2002). In addition, Runx3 RNA was detected by in situ hybridization in the stomach epithelium of type I KO mice and strong X-gal staining was observed in the stomach and intestine of E14.5 embryos (Li et al., 2002). These data do not correspond with data for the type II KO mice, in which no Runx3 expression was detected in the stomach, either by LacZ or immunostaining with specific Runx3 antibodies (Levanon et al., 2001a). The latter studies also showed that Runx1 is highly expressed in epithelia, including the gastric epithelium, whereas Runx3 expression is restricted to the mesenchymal tissues (Levanon et al., 2001a; Yamashiro et al., 2002). The genetic backgrounds of the two types of KO mice could account for the phenotypic differences observed, but what is the underlying cause of these gene expression discrepancies?
Differences in the targeting constructs
One important difference between the two mutant strains is the structure of the targeting vectors used for creating the KO mice. In type II KO mice, Runx3 was disrupted by inserting a LacZ–neomycin (neo) cassette into exon 2, the first exon of the RD (Fig. 2). Here, expression of LacZ is mediated by the distal P1 and proximal P2 promoters of Runx3, and by the IRES of the vector, thus generating a free LacZ protein (Fig. 2). In Runx3 type I KO mice, however, the LacZ–neo cassette was inserted in frame at the carboxyl terminus of the RD, creating a RD–LacZ fusion protein (Fig. 2). As the C terminus of the RD is important for DNA binding (Nagata et al., 1999; Rudolph & Gergen, 2001), the fused RD–LacZ protein would be expected to bind very poorly to DNA. However, as this fused product retains most of the RD, it may bind to the Runx partner protein CBF-β (Bravo et al., 2001) and therefore exert a dominant-negative effect (Michaud et al., 2002), particularly in the stomach of Runx3 type I KO mice where the RD–LacZ protein is highly expressed. A potential target for this negative effect is Runx1, which is expressed in the gastric mucosa of developing mouse embryos (Levanon et al., 2001a). So far, there has been no functional analysis of Runx1 activity in the stomach and intestine of either Runx3 type I or Runx3 type II KO mice, and this may prove to be informative.
Figure 2.
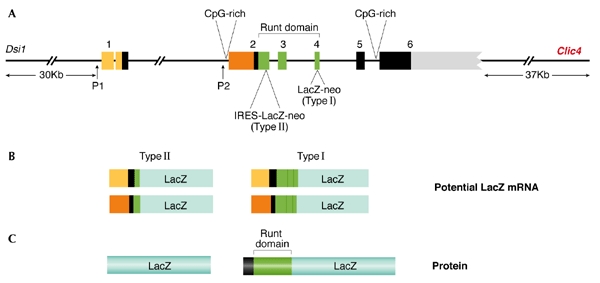
The Runx3 genomic locus including the neighbouring genes and the potential lacZ messenger RNA and protein products. (A) Genomic organization showing the two promoters (P1 and P2) and six exons. The different targeting sites in exons 2 and 4 used by Levanon et al. (2002) and Li et al. (2002), respectively and the corresponding targeting cassettes are indicated, as are the distances between Runx3 and the Dsi-1 and Clic4 loci. The positions of the CpG-rich islands at the 5′ and 3′ regions of Runx3 are marked. The various lacZ mRNAs that are transcribed from the P1 and P2 promoters in both type I and type II knockout mice are shown in (B) as are the corresponding protein products (LacZ and runt domain–LacZ) in (C). The yellow and orange segments represent the 5′ untranslated regions of P1 and P2 respectively. The green segments denote the runt domain (RD) and the blue segments represent the LacZ protein. Of note, the genomic organization of Runx3 (Bangsow et al. 2001) which is shown here including the number of Runx3 exons (1–6) differs from that in Li et al., (2002) due to the omission of exon 1 in the latter.
Structural differences within the targeted genomic locus
In addition to the structural difference in the LacZ product of the two constructs, the genomic targeting site also differs in the KO types. In type II KO mice the LacZ–neo cassette was inserted into exon 2, whereas in Runx3 type I KO mice it was placed ∼10 kb downstream, near the 3′ end of exon 4 (Fig. 2).
How might this affect the gastric phenotype? Long-distance effects on neighbouring genes are well-known phenomena. As pointed out by Balmain (2002), Runx3 is subject to upregulation in T-cell lymphomas by the insertion of murine leukemia virus (MuLV) at the Dsi1 locus, located 30 kb upstream of its P1 promoter (Stewart et al., 2002) (Fig. 2). If this, or another transposable element is present in the strain used to generate the type I KO mice, it might also affect Runx3 expression in the stomach. Interestingly, a gene that encodes the chloride intracellular channel 4 (Clic4) is located downstream of Runx3, close to the 3′-end of the gene (Figs 1, 2), and downregulation of Clic4 is known to abrogate p53-dependent apoptosis (Fernandezsalas et al., 2002). Therefore, an intriguing possibility, is that the targeting of Runx3 exon 4 might have affected the expression of Clic4 in cis, leading to attenuation of apoptosis in the gastric mucosa of Runx3 type I KO mice. In particular, the introduction of the neo gene, which is driven in the Runx3 type I KO mice by the phosphoglycerate kinase (PGK) promoter that is known to influence neighbouring genes (Scacheri et al., 2001), may have affected Clic4 expression. In this regard, it is worth noting that several Clic4 expressed sequence tags (ESTs) have previously been isolated from stomach complementary DNA libraries (for example, NCBI, UniGene Cluster Hs.25035). Analysis of Runx3 mutant mice for MuLV integration, as well as analysis after removal of the LacZ-neo cassette by Cre-recombinase–mediated excision, should help to clarify these issues.
Is the RUNX3 P1 promoter methylated in gastric cancer?
What is the connection between the downregulation of RUNX3 and human gastric cancer? Furthermore, how might the hypermethylation of the RUNX3 promoter that has been observed in primary human gastric tumours fit into the puzzle?
Gastric carcinomas, as with many other human tumours, are associated with multiple genetic alterations that affect numerous genes. These changes include genetic instability, reactivation of telomerase, inactivation of tumour-suppressor genes and activation of oncogenes (Yasui et al., 2000). Thus, it could be considered possible that epigenetic silencing of RUNX3 in human gastric tumours reflects a secondary event induced by the malignant transformation.
The regulation of RUNX3 by two promoters may also be a factor in its potential silencing, as only the P2 promoter was evaluated for hypermethylation in gastric tumours (Li et al., 2002). In contrast to the P2 promoter, which is located within a conserved CpG island (Fig. 2), the environment of the P1 promoter is CpG poor (Bangsow et al., 2001), so it is unlikely to be a target for methylation. Silencing of the P2 promoter by hypermethylation could therefore be accompanied by an upregulation of the P1 promoter (Stewart et al., 2002). Such a P2-to-P1-promoter switch has been previously observed for RUNX1 (Pozner et al., 2000). This promoter switch should not be overlooked as it could result, as with RUNX1, in an enhanced production of alternatively spliced isoforms of RUNX3 (Levanon et al., 1996), some of which could lack various parts of the carboxy-terminal region of the protein. As this missing domain is considered to be the transactivation domain of RUNX3, such isoforms may act as dominant-negative regulators. Moreover, RUNX3 isoforms that lack the trans-activation domain, but have transcription start sites at the P1 promoter, may not be detected by the RT–PCR (PCR after reverse transcription) primers used by Li et al., (2002). Examination of the methylation status of the RUNX3 P1 promoter in primary gastric tumours should shed light on this.
Perspectives
The discrepancies between the two Runx3 mutant strains will have to be reconciled through further investigation. However, it is worth noting that Runx3 KO mice have two phenotypic defects that are comparable in both type I and type II, despite variations in strain and targeting strategy. These are a prominent sensory-motor defect (Inoue et al., 2002; Levanon et al., 2002) and a well-defined defect in thymopoiesis (Taniuchi et al., 2002; Woolf et al., 2003). In both cases, Runx3 is readily detected in the affected tissues and a cell-intrinsic requirement for Runx3 has been shown. However, Runx3 has several alternatively spliced isoforms (Bangsow et al., 2001), the expression of which has not been carefully evaluated in either of the two Runx3 KO strains. Thus, the 'down but not out' phenomenon seen in other KO models (see Kos et al., 2002) should also be considered here. Furthermore, the association of Runx3 deficiency with defects in cytotoxic T-cell development (Taniuchi et al., 2002; Woolf et al. 2003) may cause secondary phenotypes that are not directly linked to Runx3 activity. Thus, the phenotypic differences described here pose the challenge of distinguishing between defects that are directly due to loss of Runx3 and those that are unrelated, non-cell-autonomous or secondary.
S.C. Bae and Y. Ito have written a response to this Concept on p538



Acknowledgments
The authors thank L. Sachs for helpful discussions and J. Lotem for comments on the manuscript. The work was supported by grants from the Commission of the European Union, the Israel Science Foundation, the Shapell Family Biomedical Research Foundation and the M.D. Moross Institute for Cancer Research at the Weizmann Institute.
References
- Adya N., Castilla L.H. & Liu P.P. (2000) Function of CBFβ/Bro proteins. Semin. Cell Dev. Biol., 11, 361–368. [DOI] [PubMed] [Google Scholar]
- Balmain A. (2002) Cancer: new-age tumour suppressors. Nature, 417, 235–237. [DOI] [PubMed] [Google Scholar]
- Bangsow C., et al. (2001) The RUNX3 gene—sequence, structure and regulated expression. Gene, 279, 221–232. [DOI] [PubMed] [Google Scholar]
- Bravo J., Li Z., Speck N.A. & Warren A.J. (2001) The leukemia-associated AML1 (Runx1)–CBFβ complex functions as a DNA-induced molecular clamp. Nature Struct. Biol., 8, 371–378. [DOI] [PubMed] [Google Scholar]
- Chen S., Gu T.T., Sreenath T., Kulkarni A.B., Karsenty G. & MacDougall M. (2002) Spatial expression of Cbfa1/Runx2 isoforms in teeth and characterization of binding sites in the DSPP gene. Connect. Tissue Res., 43, 338–344. [DOI] [PubMed] [Google Scholar]
- Eggers J.H., Stock M., Fliegauf M., Vonderstrass B. & Otto F. (2002) Genomic characterization of the RUNX2 gene of Fugu rubripes. Gene, 291, 159–167. [DOI] [PubMed] [Google Scholar]
- Fernandezsalas E. et al. (2002) mtCLIC/CLIC4, an organellular chloride channel protein, is increased by DNA damage and participates in the apoptotic response to p53. Mol. Cell. Biol., 22, 3610–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs C.S. & Mayer R.J. (1995) Gastric carcinoma. N. Engl. J. Med., 333, 32–41. [DOI] [PubMed] [Google Scholar]
- Geoffroy V., Corral D.A., Zhou L., Lee B. & Karsenty G. (1998) Genomic organization, expression of the human CBFA1 gene, and evidence for an alternative splicing event affecting protein function. Mamm. Genome, 9, 54–57. [DOI] [PubMed] [Google Scholar]
- Ghozi M.C., Bernstein Y., Negreanu V., Levanon D. & Groner Y. (1996) Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc. Natl. Acad. Sci. USA, 93, 1935–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.-H., Weng L.-Q., Ito K., Chen L.-F., Nakanishi H., Tatematsu M. & Ito Y. (2002) Inhibition of growth of mouse gastric cancer cells by Runx3 a novel tumor suppressor. Oncogene, 21, 8351–8355. [DOI] [PubMed] [Google Scholar]
- Inoue K., et al. (2002) Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nature Neurosci., 5, 946–954. [DOI] [PubMed] [Google Scholar]
- Ito Y. & Bae S.-C. (1997) in Oncogenes as Transcriptional Regulators, Vol. 2 (eds Yaniv, M. & Ghysdael, J.), 107–132. Birkhauser, Basel, Switzerland. [Google Scholar]
- Kalev-Zylinska M.L., Horsfield J.A., Flores M.V., Postlethwait J.H., Vitas M.R., Baas A.M., Crosier P.S. & Crosier K.E. (2002) Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1–CBF2T1 transgene advances a model for studies of leukemogenesis. Development, 129, 2015–2030. [DOI] [PubMed] [Google Scholar]
- Karsenty G. (2000) Role of Cbfa1 in osteoblast differentiation and function. Semin. Cell. Dev. Biol., 11, 343–346. [DOI] [PubMed] [Google Scholar]
- Kos M., Denger S., Reid G., Korach K.S. & Gannon F. (2002) Down but not out? A novel protein isoform of the estrogen receptor α is expressed in the estrogen receptor α knockout mouse. J. Mol. Endocrinol. 29, 281–286. [DOI] [PubMed] [Google Scholar]
- Kundu M. et al. (2002) Cbfβ interacts with Runx2 and has a critical role in bone development. Nature Genet., 32, 639–644. [DOI] [PubMed] [Google Scholar]
- Levanon D., Bernstein Y., Negreanu V., Ghozi M.C., Bar-Am I., Aloya R., Goldenberg D., Lotem J. & Groner Y. (1996) A large variety of alternatively spliced and differentially expressed mRNAs are encoded by the human acute myeloid leukemia gene AML1. DNA Cell Biol., 15, 175–185. [DOI] [PubMed] [Google Scholar]
- Levanon D. et al. (2002) The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. Embo J., 21, 3454–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D. et al. (2001) Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech. Dev., 109, 413–417. [DOI] [PubMed] [Google Scholar]
- Levanon D. et al. (2001) Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene, 262, 23–33. [DOI] [PubMed] [Google Scholar]
- Levanon D. et al. (2003) Phylogenesis and regulated expression of the RUNT domain transcription factors RUNX1 and RUNX3. Blood Cells Mol. Dis., 30, 161–163. [DOI] [PubMed] [Google Scholar]
- Levanon D., Negreanu V., Bernstein Y., Bar-Am I., Avivi L. & Groner Y. (1994) AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics, 23, 425–432. [DOI] [PubMed] [Google Scholar]
- Li Q.L. et al. (2002) Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell, 109, 113–124. [DOI] [PubMed] [Google Scholar]
- Lund A.H. & van Lohuizen M. (2002) RUNX: a trilogy of cancer genes. Cancer Cell, 1, 213–215. [DOI] [PubMed] [Google Scholar]
- Michaud J. et al. (2002) In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood, 99, 1364–1372. [DOI] [PubMed] [Google Scholar]
- Miller J., Horner A., Stacy T., Lowrey C., Lian J.B., Stein G., Nuckolls G.H. & Speck N.A. (2002) The core-binding factor β subunit is required for bone formation and hematopoietic maturation. Nature Genet., 32, 645–649. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Ohira M., Shimizu K., Hirai H., Imai T., Yokoyama K., Soeda E. & Ohki M. (1995) Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucl. Acids Res., 23, 2762–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T., Gupta V., Sorce D., Kim W.-Y., Sali A., Chait B.T., Shigesada K., Ito Y. & Werner M. (1999) Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nature Struct. Biol., 6, 615–619. [DOI] [PubMed] [Google Scholar]
- North T., Gu T.-L., Stacy T., Wang Q., Howard L., Binder M., Marin-Padilla M. & Speck N.A. (1999) Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development, 126, 2563–2575. [DOI] [PubMed] [Google Scholar]
- Otto F. et al. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell, 89, 765–771. [DOI] [PubMed] [Google Scholar]
- Parkin D.M., Pisani P. & Ferlay J. (1999) Estimates of the worldwide incidence of 25 major cancers in 1990. Int. J. Cancer, 80, 827–841. [DOI] [PubMed] [Google Scholar]
- Pozner A., Goldenberg D., Negreanu V., Le S.-Y., Elroystein O., Levanon D. & Groner Y. (2000) Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol. Cell. Biol., 20, 2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rini D. & Calabi F. (2001) Identification and comparative analysis of a second runx3 promoter. Gene, 273, 13–22. [DOI] [PubMed] [Google Scholar]
- Rudolph M.J. & Gergen J.P. (2001) DNA-binding by Ig-fold proteins. Nature Struct. Biol., 8, 384–386. [DOI] [PubMed] [Google Scholar]
- Scacheri P.C., Crabtree J.S., Novotny E.A., Garrett-Beal L., Chen A., Edgemon K.A., Marx S.J., Spiegel A.M., Chandrasekharappa S.C. & Collins F.S. (2001) Bidirectional transcriptional activity of PGK-neomycin and unexpected embryonic lethality in heterozygote chimeric knockout mice. Genesis, 30, 259–263. [DOI] [PubMed] [Google Scholar]
- Simeone A., Daga A. & Calabi F. (1995) Expression of runt in the mouse embryo. Dev. Dyn., 203, 61–70. [DOI] [PubMed] [Google Scholar]
- Speck N.A. (2001) Core binding factor and its role in normal hematopoietic development. Curr. Opin. Hematol., 8, 192–196. [DOI] [PubMed] [Google Scholar]
- Stewart M., MacKay N., Cameron E.R. & Neil J.C. (2002) The common retroviral insertion locus Dsi1 maps 30 kilobases upstream of the P1 promoter of the murine Runx3/Cbfa3/Aml2 gene. J. Virol., 76, 4364–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker S., Fundele R., Vortkamp A. & Mundlos S. (2002) Role of Runx genes in chondrocyte differentiation. Dev. Biol., 245, 95–108. [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.C., Komori T., Ito Y. & Littman D.R. (2002) Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell, 111, 621–633. [DOI] [PubMed] [Google Scholar]
- Wang T.C., Goldenring J.R., Dangler C., Ito S., Mueller A., Jeon W.K., Koh T.J. & Fox J.G. (1998) Mice lacking secretory phospholipase A2 show altered apoptosis and differentiation with Helicobacter felis infection. Gastroenterology, 114, 675–689. [DOI] [PubMed] [Google Scholar]
- Wheeler J.C., Shigesada K., Gergen J.P. & Ito Y. (2000) Mechanisms of transcriptional regulation by Runt domain proteins. Semin. Cell Dev. Biol., 11, 369–375. [DOI] [PubMed] [Google Scholar]
- Woolf E. et al. (2003) Runx3 and Runx1 are required for CD8 T-cell development during thymopoiesis. Proc. Natl Acad. Sci. USA, 100, (in the press.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z.S., Liu S.G., Hinson T.K. & Quarles L.D. (2001) Characterization of the upstream mouse Cbfa1/Runx2 promoter. J. Cell. Biochem., 82, 647–659. [DOI] [PubMed] [Google Scholar]
- Yamashiro T., Åberg T., Levanon D., Groner Y. & Thesleff I. (2002) Expression of Runx1,-2 and -3 during tooth, palate and cranofacial bone development. Gene Expr. Patterns., 2, 109–112. [DOI] [PubMed] [Google Scholar]
- Yasui W., Yokozaki H., Fujimoto J., Naka K., Kuniyasu H. & Tahara E. (2000) Genetic and epigenetic alterations in multistep carcinogenesis of the stomach. J. Gastroenterol., 35 (Suppl. 12.), 111–115. [PubMed] [Google Scholar]
- Yoshida C.A., Furuichi T., Fujita T., Fukuyama R., Kanatani N., Kobayashi S., Satake M., Takada K. & Komori T. (2002) Core-binding factor β interacts with Runx2 and is required for skeletal development. Nature Genet., 32, 633–638. [DOI] [PubMed] [Google Scholar]


