Abstract
Using an siRNA-based screen, we identified the transcriptional corepressor RIP140 as a negative regulator of insulin-responsive hexose uptake and oxidative metabolism in 3T3-L1 adipocytes. Affymetrix GeneChip profiling revealed that RIP140 depletion upregulates the expression of clusters of genes in the pathways of glucose uptake, glycolysis, TCA cycle, fatty acid oxidation, mitochondrial biogenesis, and oxidative phosphorylation in these cells. Conversely, we show that reexpression of RIP140 in mouse embryonic fibroblasts derived from RIP140-null mice downregulates expression of many of these same genes. Consistent with these microarray data, RIP140 gene silencing in cultured adipocytes increased both conversion of [14C]glucose to CO2 and mitochondrial oxygen consumption. RIP140-null mice, previously reported to resist weight gain on a high-fat diet, are shown here to display enhanced glucose tolerance and enhanced responsiveness to insulin compared with matched wild-type mice upon high-fat feeding. Mechanistically, RIP140 was found to require the nuclear receptor ERRα to regulate hexose uptake and mitochondrial proteins SDHB and CoxVb, although it likely acts through other nuclear receptors as well. We conclude that RIP140 is a major suppressor of adipocyte oxidative metabolism and mitochondrial biogenesis, as well as a negative regulator of whole-body glucose tolerance and energy expenditure in mice.
Introduction
Type 2 diabetes mellitus is an increasingly prevalent metabolic disorder in which the combination of systemic insulin resistance and impaired insulin production by pancreatic β cells results in hyperglycemia. Insulin regulates plasma glucose levels by selectively increasing glucose uptake into fat and muscle and by inhibiting hepatic glucose production (1). Insulin-responsive glucose uptake is initiated by a PI3K-dependent signaling cascade that stimulates trafficking of GLUT4 glucose transporters to the plasma membrane, allowing removal of excess glucose from the bloodstream (2, 3). Up to 85% of glucose disposal in humans under conditions of glucose infusion can be accounted for by insulin-stimulated glucose uptake into muscle (4), which is impaired in type 2 diabetes mellitus (5).
Enhancing fatty acid oxidation or fatty acid removal from circulation through esterification to triglyceride in adipocytes appears to correlate with increased insulin sensitivity in various mouse models (6–9). Mitochondria may be key regulators of fatty acid levels since they catalyze fatty acid oxidation as well as produce ATP, which is required for fatty acid esterification to triglyceride. Indeed, mitochondrial abundance and protein expression are increased during adipogenesis in cell culture (10). Furthermore, the insulin sensitizer rosiglitazone, a ligand for the transcription factor PPARγ, enhances mitochondrial mass and alters mitochondrial morphology in cultured adipocytes (10) and in the white adipose tissue of dogs and rats (11). Conversely, the onset of obesity and insulin resistance in ob/ob mice is accompanied by a decrease in the expression of mitochondrial genes in fat cells that is reversed by rosiglitazone treatment (12). Interestingly, microarray analysis of gene expression in human skeletal muscle revealed small decreases throughout a subset of mitochondrial genes in diabetic subjects (13, 14). Thus, mitochondrial function may be a critical regulator of glucose homeostasis in both mice and humans.
Multiple transcriptional regulators have been shown to both affect mitochondrial gene expression and to play a major role in whole-body energy balance. The PPAR family of nuclear receptors, which function as sensors for dietary lipids, play important roles in regulating lipid and carbohydrate metabolism in adipose tissue, liver, and muscle (15). The PPARγ coactivator 1α (PGC-1α) stimulates adaptive thermogenesis and mitochondrial biogenesis (16, 17), while the related coactivator, PGC-1β, regulates mitochondrial function by promoting the activity of the estrogen-related receptor α (ERRα) (18). In addition, the p160 coactivators, SRC1 and TIF2, modulate development and function of white and brown adipose tissue (19). Another important regulator is the cofactor receptor-interacting protein 140 (RIP140), a ligand-dependent corepressor for most, if not all, nuclear receptors (20, 21). It is essential for female fertility (22) and plays a crucial role in lipid metabolism (23, 24). RIP140-null mice are extremely lean and exhibit resistance to obesity and hepatic steatosis, which are normally induced in mice fed a high-fat diet (23). Adipogenesis is unaffected in these mice, but the expression of genes involved in energy dissipation and mitochondrial uncoupling, including uncoupling protein 1 (UCP1) and carnitine palmitoyltransferase 1b (CPT1b), are markedly increased in white adipose tissue. Consistent with these changes, the mice utilize more oxygen (23).
The aim of the present study was to identify novel proteins that influence insulin signaling and metabolism in adipocytes to better understand these processes. In a small interfering RNA–based (siRNA-based) screen of genes highly expressed in fat and muscle, we surprisingly identified RIP140 as a potent negative regulator of insulin-stimulated deoxyglucose uptake in 3T3-L1 adipocytes. Furthermore, RIP140 was found to regulate a broad program of metabolic gene expression by suppressing clusters of genes in the pathways of glycolysis, the TCA cycle, and mitochondrial biogenesis, in addition to its known role in regulating genes controlling fatty acid oxidation. Indeed, RIP140 depletion in cultured adipocytes increased glucose oxidation, mitochondrial protein expression, and oxygen consumption, while RIP140-null mice on a high-fat diet displayed enhanced glucose tolerance and increased responsiveness to insulin to lower blood glucose compared with wild-type mice. Thus, RIP140 functions in isolated adipocytes and in intact mice as both a major suppressor of oxidative metabolism and as a regulator of glucose homeostasis.
Results
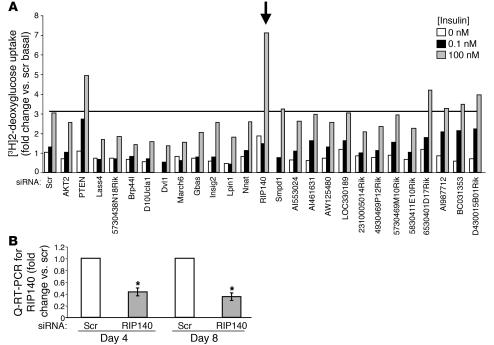
To identify novel regulators of insulin signaling and glucose uptake, we developed a miniaturized siRNA-based gene silencing screen using 3T3-L1 adipocytes. Figure 1 depicts a screen of 24 genes identified in a subtractive cDNA library that enriched transcripts highly expressed in both skeletal muscle and 3T3-L1 adipocytes compared with 3T3-L1 fibroblasts (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI26040DS1). Insulin-stimulated 2-deoxyglucose uptake was assayed 72 hours after transfection of siRNA. Figure 1A shows that depletion of the positive regulator Akt inhibited insulin stimulation of deoxyglucose uptake, as expected (25), while depletion of the known negative regulator PTEN (26) enhanced this process. While depletion of several of the candidate genes appeared to inhibit insulin action, the loss of 1 greatly enhanced insulin-responsive deoxyglucose uptake. This unique latter gene encodes the transcriptional corepressor RIP140.
Figure 1. An siRNA-mediated screen identifies Nrip1 (RIP140) as a negative regulator of deoxyglucose uptake in 3T3-L1 adipocytes.
(A) Four days after the induction of differentiation, 3T3-L1 adipocytes were transfected with smart pools of siRNA against a panel of genes (GenBank accession numbers are provided in Supplemental Table 1) selected as highly expressed in muscle and 3T3-L1 adipocytes compared with 3T3-L1 fibroblasts. The effect of each knockdown on glucose uptake was determined using a 2-deoxyglucose uptake assay. Arrow indicates insulin-stimulated 2-deoxyglucose uptake in RIP140-knockdown cells. Shown is the average of 2 independent experiments. (B) siRNA-mediated depletion of RIP140 mRNA was confirmed by real-time RT-PCR. The graph represents the mean ± SEM of 3 independent experiments. Scr, scrambled; Q-RT-PCR, quantitative RT-PCR. *P < 0.05 compared with scrambled.
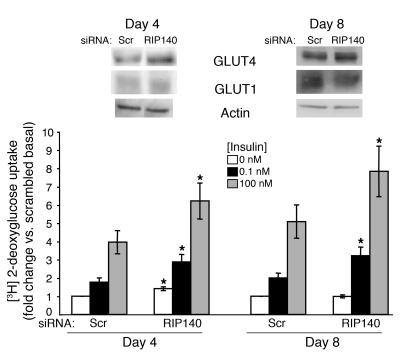
RIP140 mRNA expression is increased 4-fold during adipocyte differentiation (Supplemental Table 2 and ref. 27), and it is effectively depleted by siRNA using real-time PCR of in vitro–transcribed message (Figure 1B). Commercial antibody did not detect RIP140 in 3T3-L1 cells. RIP140 mRNA was decreased by 57% and 65%, respectively, when 3T3-L1 cells were transfected with siRNA 4 days (day 4) or 8 days (day 8) after initiation of differentiation. Differentiation of 3T3-L1 fibroblasts into adipocytes is essentially complete 8 days after its initiation (27). We found that RIP140 depletion at either point during the differentiation process enhanced insulin-stimulated deoxyglucose uptake (Figure 2), indicating that RIP140 regulates hexose uptake even in mature adipocytes. Analysis of glucose transporter proteins by Western blotting showed that RIP140 depletion increased GLUT4, but not GLUT1, protein levels approximately 1.3-fold, regardless of the day of transfection. Other indicators of differentiation, such as lipid accumulation and expression of marker genes such as aP2, were not affected by RIP140 depletion (data not shown). These results are consistent with the observed normal development of white adipose tissue in RIP140-knockout mice and the ability of RIP140-null mouse embryonic fibroblasts (MEFs) to undergo adipogenesis in culture (23).
Figure 2. RIP140 depletion enhances GLUT4 expression and hexose uptake in both day-4 and day-8 3T3-L1 adipocytes.
Four (day 4) or eight (day 8) days after the induction of differentiation, 3T3-L1 adipocytes were transfected with scrambled or RIP140 siRNA. Three days later, cell lysates were used for SDS-PAGE to assess GLUT4, GLUT1, and actin expression levels. Intact cells were used to examine the effect of RIP140 depletion on 2-deoxyglucose uptake. The graph shows the mean ± SEM of 5 independent experiments. *P < 0.05 compared with scrambled, by Student’s t test.
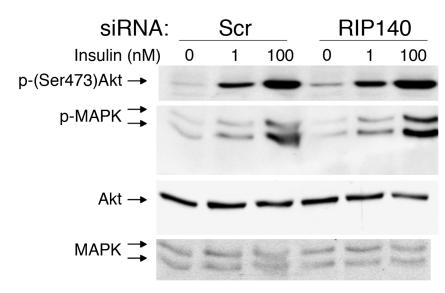
It is thought that insulin signaling to GLUT4 is mediated through activation of Akt2 (25), downstream of the PI3K signaling pathway (2). In order to test whether RIP140 might be a regulator of insulin signaling through this pathway, we tested the effect of its depletion on the rise in levels of phospho-Akt in response to insulin. No significant effect on stimulation of either Akt or MAPK due to the loss of RIP140 was observed when adipocytes were stimulated with submaximal or maximal concentrations of insulin (Figure 3). We did observe a small increase in the level of phospho-Akt in unstimulated cells in some experiments, although this effect was variable and not statistically significant.
Figure 3. Insulin signaling is not significantly affected by RIP140 silencing.
Four days after the induction of differentiation, 3T3-L1 adipocytes were transfected with scrambled or RIP140 siRNA. Three days later, the cells were starved for 2 hours and then stimulated with insulin for 30 minutes. Cell lysates were used for total and phospho-Akt and MAPK blots. The representative blot shows a small but insignificant increase in phospho-serine Akt levels.
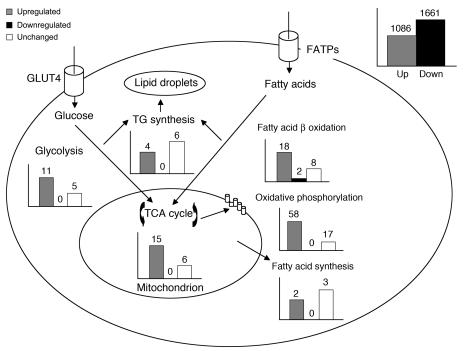
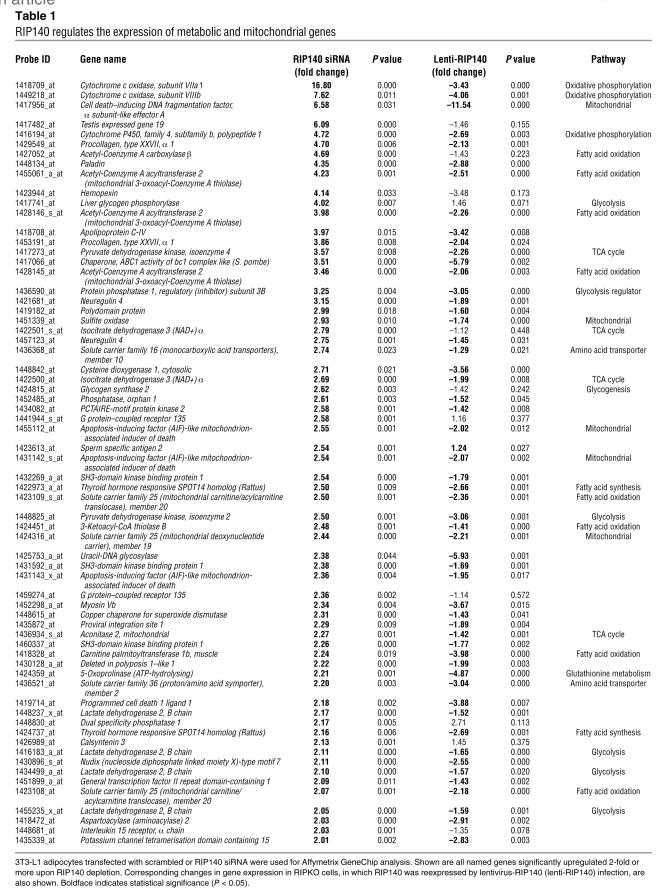
We next applied Affymetrix GeneChip expression profiling to mature 3T3-L1 adipocytes transfected with scrambled or RIP140 siRNA and analyzed the 2,747 Affymetrix probes that changed significantly (P < 0.05) upon RIP140 depletion. These genes displayed a nonrandom distribution, with a preponderance of genes encoding proteins involved in carbohydrate and fatty acid catabolic pathways (see Figure 4 and Supplemental Table 3). The majority of the genes encoding enzymes within the glycolytic, TCA cycle, fatty acid oxidative, oxidative phosphorylation, and mitochondrial biogenesis pathways were upregulated in response to RIP140 depletion (Figure 4). Mitochondrial proteins that mediate respiratory chain expression and assembly, including mitochondrial ribosomal proteins required for translation of products of the mitochondrial genome and biosynthetic enzymes required for production of cytochrome heme, were also upregulated by RIP140 depletion (data not shown). To confirm that these changes in gene expression were due directly to the loss of RIP140, we also examined changes in gene expression upon lentivirus-mediated expression of RIP140 in adipocytes derived from RIP140-null MEFs (RIPKO-1 cells described in ref. 24). Strikingly, we found a pattern of gene downregulation due to RIP140 expression that corresponded closely to the pattern of upregulation due to RIP140 depletion in 3T3-L1 adipocytes, including GLUT4 modulation (compare Figures 4 and 5, and see Supplemental Table 3). Of the top 66 genes that were significantly upregulated in RIP140-depleted 3T3-L1 adipocytes, 54 were significantly downregulated by RIP140 expression in RIPKO-1 adipocytes (Table 1). These data support a role for RIP140 as a global regulator of adipocyte genes that control metabolic and mitochondrial pathways.
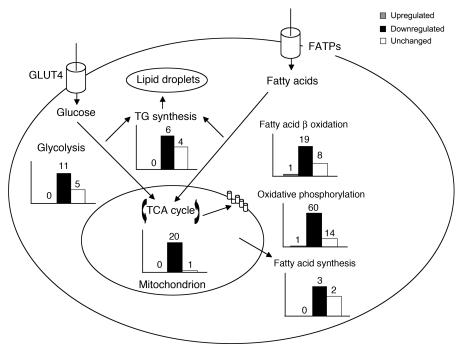
Figure 4. RIP140 silencing enhances multiple metabolic pathways in 3T3-L1 adipocytes.
Eight days after the induction of differentiation, 3T3-L1 adipocytes were transfected with scrambled or RIP140 siRNA. Three days later, mRNA was harvested and used for Affymetrix GeneChip analysis. The numbers of genes that were significantly (P < 0.05) increased, decreased, or unchanged in expression within each of the pathways shown are depicted in bar graphs adjacent to the respective pathways. The total number of significantly (P < 0.05) changed probe sets from 3 independent experiments is shown in the upper-right panel. FATP, fatty acid transport protein; TG, triglycerides.
Figure 5. RIP140 reexpression in RIPKO-1 adipocytes results in downregulation of multiple metabolic pathways.
Ten days after the induction of differentiation, RNA was isolated from RIPKO-1 adipocytes and RIPKO-1 adipocytes stably expressing RIP140 using a lentiviral vector and used for Affymetrix GeneChip analysis. The number of genes in the indicated metabolic pathways that were significantly (P < 0.05) upregulated, downregulated, or not changed is shown from left to right in the graphs.
Table 1 .
RIP140 regulates the expression of metabolic and mitochondrial genes
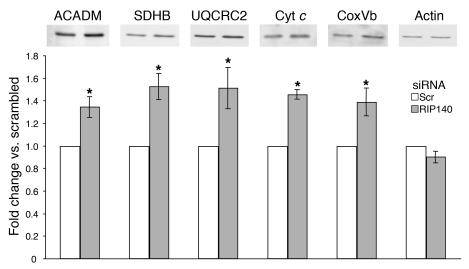
We confirmed by immunoblotting that several of the genes upregulated by RIP140 depletion showed increased expression at the protein level. The mitochondrial proteins cytochrome c (cyt c), cyt c oxidase subunit Vb (CoxVb), ubiquinol-cyt c reductase core protein 2 (UQCRC2), succinate dehydrogenase complex b (SDHB), and the fatty acid β oxidation protein medium chain acetyl-coenzyme A dehydrogenase (ACADM) were all modestly, but significantly (P < 0.05), upregulated in 3T3-L1 adipocytes upon loss of RIP140 (Figure 6).
Figure 6. Metabolic and mitochondrial protein expression is enhanced in 3T3-L1 adipocytes depleted of RIP140.
Eight days after the induction of differentiation, 3T3-L1 adipocytes were transfected with scrambled or RIP140 siRNA. After 72 hours, postnuclear supernatants were used for immunoblotting selected proteins in oxidative phosphorylation and fatty acid oxidation pathways. Shown is a representative blot and densitometric quantification of 3 independent experiments (mean ± SEM). All of the changes shown (except those for actin) were significant at P < 0.05 (indicated by asterisk).
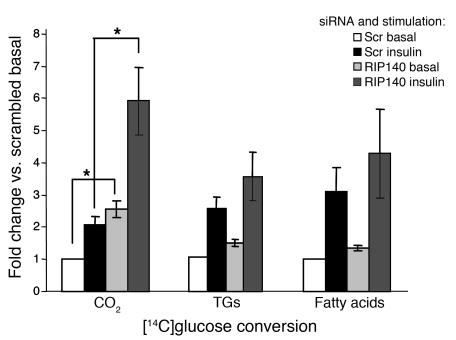
Based on the genomics analyses described above, not only GLUT4 but also hexokinase 2 and other genes within glycolytic and downstream oxidative pathways are regulated by RIP140. We therefore examined the rates of glucose oxidation, triglyceride synthesis, and de novo fatty acid synthesis in adipocytes depleted of RIP140 using d-[6-14C]-glucose. The amount of glucose converted to carbon dioxide, which is a combined measurement of glycolysis and TCA cycling, was significantly enhanced in RIP140-depleted cells in the presence or absence of insulin (Figure 7). The amount of glucose converted to triglycerides and free fatty acids, however, was not significantly affected by the loss of RIP140. These results are consistent with the gene expression profiles observed upon RIP140 depletion (Figure 4), which showed greater changes in the glycolytic (69%) and TCA (81%) pathways compared with the triglyceride synthesis (50%) or fatty acid synthesis (40%) pathways.
Figure 7. RIP140 depletion enhances glycolysis and TCA cycling, but not triglyceride synthesis or de novo free fatty acid synthesis.
Eight days after the induction of differentiation, 3T3-L1 adipocytes were transfected with scrambled or RIP140 siRNA. After 72 hours, cells were starved and glucose metabolism (with continued starvation [basal] or 1 μM insulin stimulation) was measured by [6-14C]-glucose conversion into carbon dioxide (CO2), triglycerides, and fatty acids as described in Methods. Graphs show the mean ± SEM of 3–4 independent experiments. *P < 0.05 compared with scrambled, by Student’s t test.
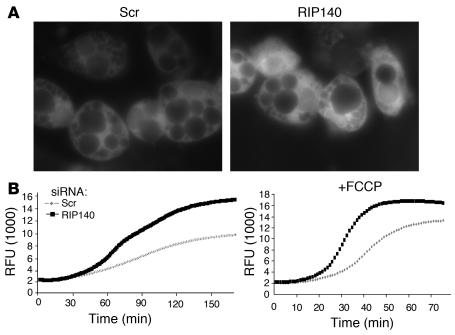
To confirm that RIP140 depletion augments mitochondrial function in cultured adipocytes, fluorescence microscopy of these cells incubated with MitoTracker Red dye was performed. Significantly increased signal was observed in RIP140-depleted cells compared with controls, indicating an increase in actively respiring mitochondria (Figure 8A). Furthermore, RIP140-depleted adipocytes consumed more oxygen in the presence or absence of the mitochondrial uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) compared with cells transfected with scrambled siRNA (Figure 8B). These data are consistent with the increased expression of genes encoding subunits within each of the respiratory complexes.
Figure 8. RIP140 is a negative regulator of cellular respiration in 3T3-L1 adipocytes.
Eight days after the induction of differentiation, 3T3-L1 adipocytes were transfected with scrambled or RIP140 siRNA. (A) After 72 hours, cells were incubated with MitoTracker Red to stain actively respiring mitochondria. Fluorescence microscopy was performed as described in Methods. Shown are representative photographs with identical exposure times. (B) Cells were assayed for respiration using the oxygen biosensor fluorescence assay described in Methods. Basal respiration is indicated on the left, and uncoupled respiration, induced by addition of the mitochondrial uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP; 33 μM), is shown on the right. Illustrated are the averages of 3–4 independent experiments. The respiration with or without FCCP of cells transfected with scrambled or RIP140 siRNA are considered statistically different based on the fluorescence intensity at the time when fluorescence of the leading curve was equal to half-maximum for each experiment. RFU, raw fluorescence units.
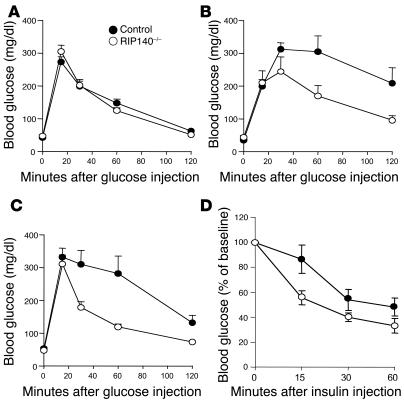
Because increased mitochondrial function is correlated with improved glucose metabolism, RIP140-null mice (23) were subjected to glucose tolerance tests. As shown in Figure 9A, young wild-type mice and RIP140-knockout mice fed a normal chow diet displayed similar increases in blood glucose concentrations after injection of a glucose load. In contrast, RIP140-null mice exhibited significantly improved glucose tolerance at 8 months of age (Figure 9B) or after a high-fat diet (Figure 9C) compared with wild-type mice matched for age and diet. We also tested whether insulin sensitivity is affected by RIP140 expression levels in mice. An insulin tolerance test demonstrated that RIP140-null mice have increased insulin responsiveness with respect to glucose lowering when on a high-fat diet (Figure 9D). These data indicate that RIP140-null mice show both higher insulin sensitivity and glucose tolerance under circumstances that normally inhibit these processes in wild-type mice. This phenotype is consistent with results described above in cultured adipocytes showing that RIP140 is a negative regulator of oxidative metabolism, although more work is needed to determine whether the role of RIP140 in adipocytes is sufficient to explain the whole-body effects.
Figure 9. Absence of RIP140 protects against age- and diet-induced glucose intolerance and enhances insulin responsiveness on a high-fat diet.
Glucose tolerance tests were performed on 3-month-old female mice fed a normal chow diet (A), 8-month-old female mice fed a normal chow diet (B), or 3-month-old female mice fed a high-fat diet (C) for 10 weeks. Values are mean ± SEM; n = 6–12. (D) Insulin tolerance tests were performed on 3-month-old female mice fed a high-fat diet for 9 weeks. Values are mean ± SEM; n = 9.
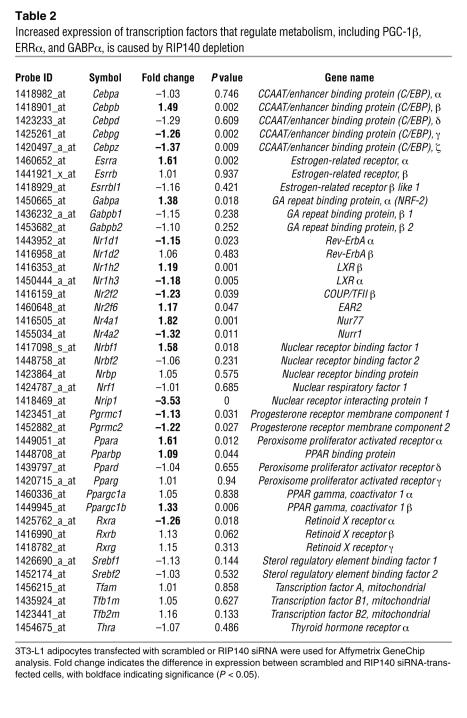
RIP140 could regulate the expression of metabolic genes by repressing nuclear hormone receptors that act directly at the promoters of these genes. Alternatively, RIP140 could repress nuclear receptors that act indirectly by regulating expression levels of transcription factors that in turn regulate metabolic gene promoters. To evaluate the latter possibility, we examined the microarray-based gene expression database described above for expression profiles of transcription factors and cofactors known to regulate genes encoding mitochondrial proteins. The PPARγ coactivator PGC-1α, which has been shown to regulate mitochondrial biogenesis in brown adipose tissue, muscle, and liver (16, 28), is expressed at very low levels in white adipose tissue and 3T3-L1 adipocytes, and its expression did not change significantly upon RIP140 depletion. Expression of nuclear respiratory factor 1 (28) and thyroid hormone receptor (29), both reported to be important in mitochondrial biogenesis, was unchanged as well. However, a modest but significant increase in the expression of the PPARγ coactivator PGC-1β in RIP140-depleted cells was evident (Table 2). PGC-1β is known to enhance expression of genes related to energy metabolism (18, 30). Furthermore, increases in the expression of ERRα and GA repeat-binding protein α (GABPα), which have been reported to act downstream of PGC-1α to regulate oxidative phosphorylation in skeletal muscle (31, 32), were observed (Table 2). Finally, the absence of changes in expression of Tfam, Tfb1, and Tfb2 might reflect the absence of increased transcription or replication of the mitochondrial genome. However, the elevated levels of mitochondrial ribosomal proteins observed in RIP140-depleted cells might increase translation of the 13 subunits of the respiratory chain encoded in mitochondrial DNA. Taken together, these results indicate that RIP140 suppresses the expression of several positive regulators of mitochondrial protein expression, providing 1 potential mechanism for its action to increase mitochondrial protein levels.
Table 2 .
Increased expression of transcription factors that regulate metabolism, including PGC-1β, ERRα, and GABPα, is caused by RIP140 depletion
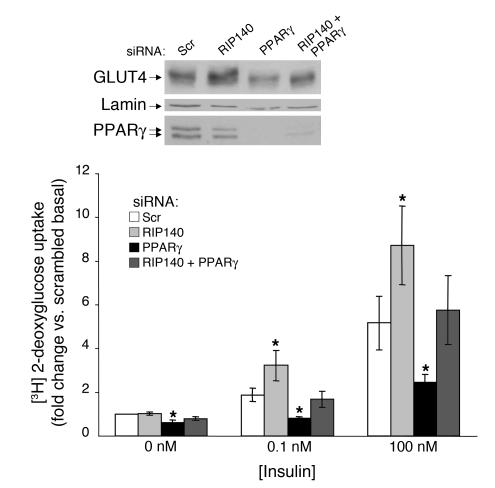
The enhancement in insulin-responsive deoxyglucose uptake and mitochondrial protein expression caused by RIP140 depletion resembles the increased insulin sensitivity (33), glucose uptake (34–36), and mitochondrial biogenesis (10) elicited by treatment of adipocytes with rosiglitazone, a PPARγ agonist. RIP140 was shown to interact with PPARγ in a yeast 2-hybrid assay and in vitro (37). To investigate whether RIP140 regulates GLUT4 expression and glucose uptake by modulating PPARγ activity, we used siRNA to deplete RIP140, PPARγ, or both proteins in fully differentiated 3T3-L1 adipocytes. As expected, depletion of RIP140 enhanced GLUT4 expression and deoxyglucose uptake (Figure 10). PPARγ depletion resulted in reduced GLUT4 levels, which is consistent with the requirement of PPARγ for the maintenance of the adipocyte phenotype (38). Interestingly, depletion of RIP140 in combination with PPARγ depletion caused an enhancement of GLUT4 expression and deoxyglucose uptake compared with PPARγ depletion alone. This suggests that RIP140 may be able to regulate GLUT4 expression independently of PPARγ and that other nuclear receptors are involved.
Figure 10. RIP140 depletion does not require normal PPARγ levels to enhance GLUT4 expression.
Eight days after the induction of differentiation, cells were transfected with scrambled, RIP140, PPARγ, or RIP140 plus PPARγ siRNA. Three days later, cell lysates were used for immunoblotting GLUT4 and actin, while nuclear extracts were used to immunoblot PPARγ and lamin. A 2-deoxyglucose uptake assay was performed with the indicated knockdowns. The graph shows the average and SE of 5 independent experiments. *P < 0.05 compared with scrambled siRNA, by Student’s t test.
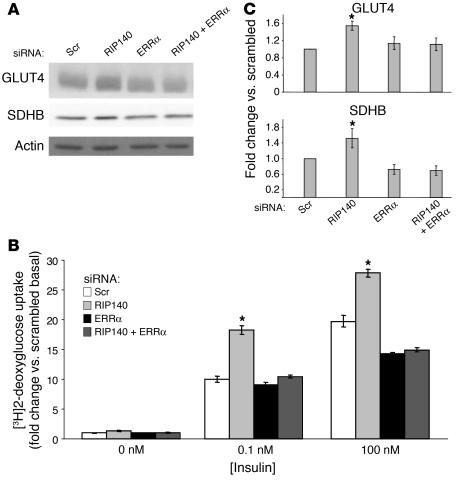
We thus investigated whether RIP140 uses an ERRα-dependent mechanism to regulate GLUT4 and mitochondrial gene expression, as ERRα is thought to be an important regulator of energy balance (39) and oxidative phosphorylation–related gene expression (32) and its expression is increased in RIP140-depleted adipocytes (Table 2). ERRα depletion alone in 3T3-L1 adipocytes modestly decreased GLUT4 and SDHB levels and inhibited deoxyglucose uptake compared with cells treated with scrambled siRNA (Figure 11). Strikingly, the loss of ERRα prevented the increased GLUT4 and SDHB expression and deoxyglucose uptake due to RIP140 depletion. Similar results were seen with the mitochondrial protein CoxVb (data not shown). These data suggest that RIP140 regulates GLUT4 and mitochondrial gene expression by a mechanism that is dependent upon ERRα.
Figure 11. RIP140 depletion requires ERRα to enhance GLUT4 expression, deoxyglucose uptake, and mitochondrial protein expression.
Eight days after the induction of differentiation, cells were transfected with scrambled, RIP140, ERRα, or RIP140 plus ERRα siRNA. (A) Three days later, cell lysates were used for immunoblotting GLUT4, SDHB, and actin. A representative blot and the mean ± SEM of 4–5 independent experiments are shown. (B) A 2-deoxyglucose uptake assay was performed with the indicated knockdowns. A representative blot and the mean ± SEM of 3 independent experiments are shown. *P < 0.05 compared with scrambled siRNA, by Student’s t test.
Discussion
The findings presented here demonstrate that the transcriptional corepressor RIP140 negatively regulates major gene clusters in the pathways of carbohydrate and fatty acid catabolism and mitochondrial biogenesis in cultured adipocytes. Depletion of RIP140 in 3T3-L1 adipocytes caused increased expression of clusters of genes in these metabolic pathways, whereas its reintroduction into RIPKO-1 adipocytes caused decreased expression of these same genes (Figures 4 and 5 and Supplemental Table 3). Expression of large numbers of genes encoding proteins that function in all of the mitochondrial electron transport complexes was increased by RIP140 loss in adipocytes and resulted in an increase in overall mitochondrial functioning (Figures 7 and 8). Specifically, RIP140 suppression enhanced hexose uptake (Figures 1 and 2) and conversion of labeled glucose to CO2 (Figure 7) and markedly elevated mitochondrial oxygen consumption in cultured adipocytes (Figure 8). Our results also agree with those of Leonardsson et al. (23) and Soukas et al. (27), demonstrating that RIP140 expression is markedly increased during adipocyte differentiation from preadipocytes (Supplemental Table 2). Taken together, these findings indicate that in adipocytes, the highly expressed protein RIP140 exerts a major suppressive effect on oxidative metabolism in the glycolytic and TCA cycle pathways as well as mitochondrial biogenesis, in addition to its known role in fatty acid oxidation.
The dramatic effect of RIP140 depletion in upregulating genes encoding metabolic enzymes and proteins that catalyze mitochondrial respiration in adipocytes raised interesting questions about the potential consequences of RIP140 deletion on whole-body glucose homeostasis. Adipogenesis itself enhances insulin sensitivity of glucose utilization as well as mitochondrial gene expression (10), both of which are expected to enhance triglyceride synthesis. Lipogenesis in adipocytes in turn is thought to be 1 pathway whereby fatty acids are sequestered away from other tissues such as muscle, reducing insulin resistance (33). Interestingly, obesity and insulin resistance in ob/ob mice are accompanied by a decrease in mitochondrial gene expression in white adipose tissue (12). Long-term calorie restriction, which results in enhanced insulin sensitivity, causes an increase in the expression of mitochondrial genes in white adipose tissue (40). Also, subcutaneous fat, which is more insulin sensitive than visceral fat, has a higher mitochondrial density, mitochondrial DNA content, and mitochondrial activity than visceral fat (41). Furthermore, rosiglitazone treatment of ob/ob mice enhances expression of genes encoding mitochondrial proteins in adipose tissue while at the same time enhancing muscle insulin sensitivity and glucose tolerance (10). These data, coupled with results from experiments showing inverse relationships between the amount of GLUT4 glucose transporters in adipocytes and whole-body insulin resistance (4, 42, 43), reinforce the hypothesis that oxidative metabolism in adipocytes may indirectly control muscle insulin sensitivity and whole-body glucose homeostasis.
The above considerations prompted us to test whether RIP140-null mice might exhibit increased glucose tolerance and insulin sensitivity. RIP140-null mice were previously shown to be lean and resistant to high-fat diet–induced obesity (23). However, these mice did not initially show a change in glucose tolerance at early ages on a normal diet. Here we show that RIP140-null mice are resistant to the impaired glucose tolerance and poor insulin responsiveness observed in wild-type mice fed a high-fat diet (Figure 9, C and D). Enhanced glucose tolerance in aged RIP140-null mice was also observed compared with matched wild-type mice (Figure 9B). Taken together, these data indicate that the increased glucose tolerance observed in RIP140-null mice on a high-fat diet is due to increased insulin responsiveness. Thus, RIP140 may be an important modulator of potential linkages between adipocyte metabolism and whole-body glucose tolerance. Our data show only a correlation between these processes at this stage, which should provoke further work to rigorously test this hypothesis.
The expression of genes encoding mitochondrial proteins in skeletal muscle has recently been correlated with insulin sensitivity in humans (44). Many studies focused on skeletal muscle have indicated the importance of mitochondrial size and number (45, 46) and expression of genes encoding proteins that catalyze oxidative phosphorylation and mitochondrial capacity (13, 14, 47–50). A recent study also found that depletion of mitochondrial DNA leads to a dramatic decrease in glucose uptake in skeletal muscle (51). It will thus be interesting to determine whether RIP140 modulates energy metabolism in skeletal muscle, as it does in fat. The degree to which RIP140 depletion in muscle or other tissues contributes to the enhanced glucose tolerance and insulin sensitivity of RIP140-null mice is an important question and remains to be determined.
While our data show that many proteins of the mitochondrial electron transport chain and oxidative phosphorylation are negatively controlled by RIP140, the expression of mitochondrial uncoupling proteins was not apparently modulated by RIP140 depletion in 3T3-L1 adipocytes (data not shown). In contrast, Leonardsson et al. did observe expression of the uncoupling protein UCP1 in white adipose tissue of RIP140-null mice (23), a protein normally expressed only in brown fat, as did Christian et al. in differentiated RIP140-null MEFs (24). This discrepancy might be due to the presence of RIP140 in 3T3-L1 adipocytes prior to its depletion by siRNA and during a critical time period when UCP1 expression is determined, whereas in RIP140-null mice and MEFs, RIP140 was never present. UCP1 is thought to divert the energy derived from mitochondrial electron transport that would normally generate ATP to produce heat. This uncoupling effect greatly accelerates oxygen consumption. In intact mice, RIP140 might act to divert white adipocytes from a brown fat phenotype by attenuating the expression of UCP1 and genes involved in fatty acid oxidation and mitochondrial respiration, all of which are abundantly expressed in brown fat. Interestingly, conversion of white adipocytes to a brown fat phenotype has been achieved by ectopic expression of PGC-1α in human adipocytes (52), suggesting that RIP140 and the PGC family coactivators may have antagonizing roles in adipocyte development.
RIP140 appears to interact with multiple nuclear receptors through its LXXLL motifs, including PPARγ (37), which promotes and maintains the adipocyte phenotype (53). Both RIP140 depletion and the treatment of 3T3-L1 adipocytes or obese ob/ob mice with the PPARγ agonist rosiglitazone cause an increase in the expression of genes encoding mitochondrial proteins (12). We therefore tested whether RIP140 might act by a mechanism involving direct repression of PPARγ to regulate glucose uptake and GLUT4 expression. PPARγ depletion in mature adipocytes by siRNA transfection did decrease deoxyglucose transport and GLUT4 expression, as expected. However, the loss of RIP140 in these cells was still able to enhance these processes, even with greatly reduced amounts of PPARγ (Figure 10). Taken together, these data indicate that PPARγ is either not a major target of RIP140-mediated repression responsible for regulating GLUT4 expression or that other pathways can also mediate this response in parallel. Further experiments will be required to determine whether other genes responsive to RIP140 involve PPARγ regulation by the corepressor.
RIP140 depletion may increase gene expression in cultured adipocytes by upregulating the expression of ERRα (Table 2). We found that RIP140 knockdown requires the presence of ERRα to enhance GLUT4 expression and deoxyglucose uptake (Figure 11), suggesting a potential mechanism in which RIP140 represses ERRα-mediated transcription of GLUT4 directly or indirectly through other regulatory genes. ERRα is also required for RIP140 regulation of other genes, including oxidative phosphorylation proteins such as CoxVb and SDHB. Interestingly, expression of a transcriptionally active ERRα was sufficient to induce mitochondrial biogenesis in an osteoblast cell line with adipocyte differentiation capacity (31). Thus, RIP140 depletion may activate ERRα, leading to an increase in mitochondrial content and GLUT4 expression. The activation of ERRα might also play a role in the increases in expression of genes that mediate fatty acid oxidation (18), although further work is needed to test this hypothesis.
The repression of genes related to oxidative phosphorylation and fatty acid oxidation by RIP140 is likely to involve additional factors. In skeletal muscle, for example, the coactivator PGC-1α induces ERRα and GABPα, which then turn on the transcription of genes that encode a subset of the proteins that mediate oxidative phosphorylation (32). Thus, RIP140 depletion may require GABPα and the related coactivator PGC-1β, a coactivator for ERRα (18), to enhance oxidative phosphorylation gene expression. Similarly, PPARα may be required for ERRα-induced increases in fatty acid oxidation (54), although both PGC-1β (55, 56) and ERRα (54) have been shown to activate PPARα. Interestingly, each of these additional regulators (GABPα, PGC-1β, and PPARα) exhibit increased expression upon RIP140 depletion (Table 2). Finally, several other transcription factors are increased in RIP140-depleted adipocytes and may contribute to the enhanced insulin sensitivity and metabolism of these cells.
In summary, we have shown here that the metabolic pathways suppressed by RIP140 in adipocytes include glycolysis, triglyceride synthesis, the TCA cycle, fatty acid β oxidation, mitochondrial electron transport, and oxidative phosphorylation. Indeed, glucose oxidation and oxygen consumption are significantly increased in adipocytes when RIP140 function is attenuated by siRNA silencing. RIP140 regulates GLUT4 and mitochondrial protein expression by a mechanism that is dependent upon ERRα in cultured adipocytes, although GLUT4 is not regulated in the adipose tissue of RIP140-null mice. Finally, RIP140-null mice display enhanced glucose and insulin tolerance after high-fat feeding compared with matched wild-type mice. Since RIP140 exerts broad negative regulation of multiple metabolic pathways in adipocytes, it may in part mediate changes in adipocyte metabolism in response to a variety of physiological conditions in order to effectively alter whole-body glucose metabolism. Furthermore, our results suggest that RIP140 may be a candidate therapeutic target for the syndromes of obesity and type 2 diabetes.
Methods
Materials and chemicals.
Human insulin was obtained from Eli Lilly and Co. Rabbit polyclonal antibodies against phospho-Akt (Ser473), phospho-Erk1/2, anti-Akt, and anti-Erk1/2 were from Cell Signaling Technology. Other antibodies included goat anti-GLUT4 (Santa Cruz Biotechnology Inc.), mouse anti-actin (Sigma-Aldrich), rabbit anti-GLUT1 (gift from Paul Pilch, Boston University, Boston, Massachusetts, USA), mouse anti–cyt c (BD Biosciences — Pharmingen), and mouse anti-CoxVb (MitoSciences). Goat anti-ACADM, mouse anti-UQCRC2, and mouse anti-SDHB antibodies were from Abcam. The HRP-conjugated goat anti-rabbit IgG and rabbit anti-goat IgG were from Chemicon International.
Subtractive cDNA library.
The Muscle/Adipocyte Union cDNA library was made using a modification of the subtractive suppression hybridization technique of the Clontech PCR-Select cDNA Subtraction Kit (57).
siRNA.
siRNA was purchased from Dharmacon. Smart pools of siRNA consisting of a mix of 4 individual oligonucleotides were used for the screen and to deplete ERRα. Individual siRNA sequences included: scrambled, 5′-CAGTCGCGTTTGCGACTGG-3′; RIP140, 5′-GGAATGAGCTCGATTATAA-3′; and PPARγ, 5′-GACATGAATTCCTTAATGA-3′. Smart pool and individual siRNA-mediated depletion of RIP140 and PPARγ produced similar experimental results.
Cell culture and electroporation of 3T3-L1 adipocytes.
3T3-L1 fibroblasts and RIP140-null MEFs, referred to as RIPKO cells (24), were grown in DMEM supplemented with 10% FBS, 50 μg/ml streptomycin, and 50 U/ml penicillin and differentiated into adipocytes as described previously (25). 3T3-L1 adipocytes were transfected with siRNA duplexes by electroporation on the fourth or eighth day following the induction of differentiation (day 4 or day 8) as described previously (25). The efficiency of siRNA electroporation using this method is greater than 95%, based on fluorescence microscopy of cells electroporated with Cy3-siRNA.
2-Deoxyglucose uptake assay.
Insulin-stimulated glucose transport in 3T3-L1 adipocytes was estimated by measuring 2-deoxyglucose uptake using a miniaturized version of the typical assay with cells seeded into 96-well plates (25). Cells were solubilized with 0.1 ml of 1% SDS, and 3H uptake was quantitated by scintillation counting. Nonspecific deoxyglucose uptake was measured in the presence of 20 μM cytochalasin B and subtracted from each determination to obtain specific uptake.
Quantitative RT-PCR.
Adipocytes were transfected as described above. Total RNA was prepared 24 hours after transfection using TRIzol reagent (Invitrogen Corp.). cDNA was synthesized using oligo-dT primers and AMV reverse transcriptase (Roche Diagnostics Corp.) according to manufacturer’s instructions. Quantitative real-time PCR was performed in a LightCycler (Roche Diagnostics Corp.) using a SYBR Green I PCR kit (Roche Diagnostics Corp.) and specific primers to amplify a 553-bp fragment from RIP140 or a 500-bp fragment from hypoxanthine phosphoribosyl transferase.
Immunoblotting.
Cell lysates were harvested by the addition of SDS lysis buffer (2% SDS, 30 mM NaCl, 10 mM HEPES, pH 7.4, 20 mM NaF, 1 mM NaPPi). For detecting phospho-proteins, cells were starved overnight in serum-free DMEM medium with 0.5% BSA. Cells were then incubated with 0, 1, or 100 μM insulin for 30 minutes and harvested with 1% SDS lysis buffer as described above. For enrichment of nuclear proteins (PPARγ, lamin A/C blots), cells were lysed in hypotonic lysis buffer (20 mM Tris-HCl, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 1 mM DTT, 1 mM sodium orthovanadate, 30 mM β-glycerophosphate, 1.5% NP-40) using a Dounce homogenizer. Nuclei were pelleted (3,500 g for 5 minutes at 4°C), washed in hypotonic lysis buffer, and pelleted again. They then were resuspended in NUN buffer (1.1 M urea, 330 mM NaCl, 1.1% NP-40, 17.5 mM HEPES, pH 7.6, 100 mM DTT, 100 mM sodium orthovanadate, 3 M β-glycerophosphate) and incubated on ice for 30 minutes. A final spin at 16,000 g for 10 minutes at 4°C was performed, and the extracted nuclear proteins in the supernatant were mixed with glycerol and stored at –80°C. Postnuclear supernatants were used for immunoblotting ACADM, cyt c, CoxVb, UQCRC2, and SDHB. Equal amounts of protein from lysates were resolved by SDS-PAGE, immunoblotted, and detected with an enhanced chemiluminescence kit (PerkinElmer).
Affymetrix GeneChip analysis.
Isolation of RNA and Affymetrix GeneChip analysis were performed as described for fibroblasts, 3T3-L1 adipocytes, and primary fat cells (10). For RIP140 3T3-L1 Affymetrix GeneChip analysis, cells were transfected as described above. RNA was prepared from TRIzol following manufacturer’s instructions, and cDNA and cRNA were made using Affymetrix’s One-Cycle cDNA Synthesis and IVT Labeling Kits. cRNA was hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 array. All microarray experiments were performed in triplicate using separate plates of cells.
3T3-L1 GeneChip data was analyzed using the robust multiarray average algorithm as a function of the BioConductor Affy R package. The preprocessed data is stored as log2-transformed real signal numbers. The fold change of expression of a gene in 2 experiments is the ratio of mean signal values from these experiments. To determine differential expression of genes in 2 hybridization experiments, we conducted a 2-tailed Student’s t test with the expression signal values of the 2 hybridizations for all genes in the set. The P values were not adjusted for multiple testing. P < 0.05 was considered statistically significant.
For Affymetrix analysis of gene expression in MEFs derived from RIP140-null mice, with or without RIP140 lentiviral reexpression, total RNA was isolated from 3 samples each after adipocyte differentiation (10 days) in the presence of the PPARγ ligand GW1929 (2.5 μm; Sigma-Aldrich). Two RIP140-lentivirus transduced cell lines were used for profiling in order to avoid variation due to different sites of incorporation of the virus. Affymetrix array hybridization and scanning were performed by the CSC/IC Microarray Centre, Imperial College London, Hammersmith Campus, using Murine 430 2.0 chips. Array data were analyzed with d-CHIP software and are available at http://www.ebi.ac.uk/arrayexpress/, accession number E-MIMR-42.
Tables list only genes indicated as present by Affymetrix, except for Supplemental Table 2. Only 1 representative probe set per gene is shown if more than 1 probe set was present. Boldface in Tables 1 and 2 and shading in supplemental tables indicate that gene expression was significantly changed, with P < 0.05.
Glucose conversion to CO2, triglycerides, and free fatty acids.
Adipocytes were transfected as described above and reseeded into 15-cm dishes. Cells were washed twice and starved for 2 hours with Krebs-Ringer–HEPES buffer (KRH buffer; 130 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 1.3 mM MgSO4, 25 mM HEPES, pH 7.4) supplemented with 2.5% BSA and 2 mm sodium pyruvate. Detached cells were aliquoted in triplicate to tubes containing supplemented KRH buffer (basal), 1 μM insulin, or 20 μM cytochalasin B, and d-[6-14C]-glucose was added. After 2 hours at 37°C, [3H]CO2 was released by the addition of 2 M HCl, and collection filter papers were scintillation counted to measure glucose conversion to CO2. Five milliliters of Dole’s solution (80 ml isopropanol, 20 ml hexane, 2 ml 0.5 M H2SO4) was added and incubated for 15 minutes. Hexane was used to extract the lipids, and 1 ml was transferred to a scintillation vial, evaporated, and used to measure glucose conversion to triglycerides. Another 1 ml of the hexane phase was evaporated, and 1 ml of ethanolic KOH was added. Vials were heated at 80°C for 1 hour to release fatty acids from triglycerides and washed with water, and the pH was adjusted with 3 M sulfuric acid. Hexane was added, evaporated, and used to measure glucose conversion to fatty acids. DNA content was assessed using the Qiagen DNeasy kit, and counts per minute were converted to picomoles glucose and normalized to the number of cells per tube.
Oxygen consumption assays.
Three days following transfection with siRNA, adipocytes were resuspended in KRH buffer supplemented with 2.5% BSA and 2 mm sodium pyruvate. Equal numbers of cells were seeded in triplicate into BD Oxygen Biosensor System plates (BD Biosciences). Plates were pre-read to determine baseline fluorescence and were read on a Safire multimode microplate spectrophotometer (Tecan) at 1-minute intervals for 150 minutes. An excitation wavelength of 485 nm and emission wavelength of 630 nm were used. The mitochondrial uncoupler FCCP (Sigma-Aldrich) was used at a final concentration of 33 μM. Data were analyzed by averaging the triplicates in each experiment and determining the maximum fluorescence value for the leading curve. Fluorescence intensities were collected at the time point corresponding to half-maximal fluorescence of the leading curve.
Fluorescence microscopy.
Adipocytes transfected with siRNA were seeded directly onto coverslips. After 3 days, mitochondrial respiration was measured by incorporation of 1 μM MitoTracker Red (Molecular Probes; Invitrogen Corp.) for 30 minutes at 37°C. Cells were washed and fixed for 10 minutes using 4% formaldehyde. Coverslips were mounted onto DABCO on slides and sealed. Imaging was performed using a wide-field microscope with a ×100 Nikon PlanApo objective. Image collection and quantitation were performedt blindly. The total fluorescence intensity of a minimum of 50 cells was determined for each experiment by outlining each cell in Adobe Photoshop CS, version 8.0 (Adobe Systems Inc.) and multiplying the number of pixels by the average pixel intensity from the histogram function.
RIP140-null mice and glucose and insulin tolerance tests.
RIP140-null mice were generated as previously described (22). Mice used in this study were backcrossed 8 generations to a C57BL/6J background. Mice were maintained under standard conditions, with controlled light and temperature, and fed a chow diet ad libitum, except in high-fat diet experiments, wherein mice were fed a 45% kcal fat diet (Research Diets). All animal work was performed with approval of the Home Office (London, United Kingdom). Glucose and insulin tolerance tests were performed on mice after a 16-hour overnight fast. Animals were injected intraperitoneally with d-glucose (2 g/kg) or insulin and blood glucose levels measured at the time points indicated. Blood glucose was measured using a Glucometer Elite (Bayer Corp).
Supplementary Material
Acknowledgments
We appreciate many helpful discussions with Jennifer Christianson, Alison Burkart, My Chouinard, and Silvia Corvera. Many thanks go to Jane Erickson for assistance with the manuscript. This work was supported by NIH grants DK30898 to M.P. Czech, D32520-20 to J. Straubhaar, and DK60837-03 to the Diabetes Genome Anatomy Project; CytRx Corp.; and Wellcome Trust grant 061930 to A. Seth and E. Kiskinis. We are also grateful to the staff of the CSC/Imperial College Microarray Centre for assistance with the microarray hybridization, staining, and data analysis.
Footnotes
Nonstandard abbreviations used: ACADM, medium chain acetyl-Coenzyme A dehydrogenase; Cox Vb, cyt c oxidase subunit Vb; cyt c, cytochrome c; ERRα, estrogen related receptor α; GABPα, GA repeat-binding protein α; KRH, Krebs-Ringer–HEPES; MEF, mouse embryonic fibroblast; PGC-1α, PPARγ coactivator 1α; RIP140, receptor-interacting protein 140; SDHB, succinate dehydrogenase complex b; siRNA, small interfering RNA; UCP1, uncoupling protein 1; UQCRC2, ubiquinol-cyt c reductase core protein 2.
Conflict of interest: M.G. Parker is entitled to a share of revenue under Imperial College’s discretionary rewards to inventors scheme. Imperial College London has received equity and may receive future consideration from CytRx Corp. under an existing license agreement. M.P. Czech, A.M. Powelka, X. Tang, A.D. Cherniack, and A. Guilherme own, or may be provided, equity in CytRx Inc., based on intellectual property licensed from the University of Massachusetts Medical School, which shares royalties with inventors. M.P. Czech is a consultant for CytRx Inc.
Citation for this article: J. Clin. Invest. 116:125–136 (2006).doi:10.1172/JCI26040
Andrew D. Cherniack’s present address is: Abbott Laboratories, Worcester, Massachusetts, USA.
References
- 1.Saltiel A.R., Kahn C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 2.Czech M.P., Corvera S. Signaling mechanisms that regulate glucose transport. J. Biol. Chem. 1999;274:1865–1868. doi: 10.1074/jbc.274.4.1865. [DOI] [PubMed] [Google Scholar]
- 3.Watson R.T., Pessin J.E. Subcellular compartmentalization and trafficking of the insulin-responsive glucose transporter, GLUT4. Exp. Cell. Res. 2001;271:75–83. doi: 10.1006/excr.2001.5375. [DOI] [PubMed] [Google Scholar]
- 4.Minokoshi Y., Kahn C.R., Kahn B.B. Tissue-specific ablation of the GLUT4 glucose transporter or the insulin receptor challenges assumptions about insulin action and glucose homeostasis. J. Biol. Chem. 2003;278:33609–33612. doi: 10.1074/jbc.R300019200. [DOI] [PubMed] [Google Scholar]
- 5.Cline G.W., et al. Impaired glucose transport as a cause of decreased insulin-stimulated muscle glycogen synthesis in type 2 diabetes. N. Engl. J. Med. 1999;341:240–246. doi: 10.1056/NEJM199907223410404. [DOI] [PubMed] [Google Scholar]
- 6.Yamauchi T., et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 2002;8:1288–1295. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- 7.Ntambi J.M., et al. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11482–11486. doi: 10.1073/pnas.132384699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh W., et al. Glucose and fat metabolism in adipose tissue of acetyl-CoA carboxylase 2 knockout mice. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1384–1389. doi: 10.1073/pnas.0409451102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joseph J.W., et al. Free fatty acid-induced beta-cell defects are dependent on uncoupling protein 2 expression. J. Biol. Chem. 2004;279:51049–51056. doi: 10.1074/jbc.M409189200. [DOI] [PubMed] [Google Scholar]
- 10.Wilson-Fritch L., et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toseland C.D., Campbell S., Francis I., Bugelski P.J., Mehdi N. Comparison of adipose tissue changes following administration of rosiglitazone in the dog and rat. Diabetes Obes. Metab. 2001;3:163–170. doi: 10.1046/j.1463-1326.2001.00117.x. [DOI] [PubMed] [Google Scholar]
- 12.Wilson-Fritch L., et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Invest. 2004;114:1281–1289. doi: 10.1172/JCI200421752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mootha V.K., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 14.Patti M.E., et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans R.M., Barish G.D., Wang Y.X. PPARs and the complex journey to obesity. Nat. Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 16.Puigserver P., Spiegelman B.M. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 17.Puigserver P., et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/S0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 18.Kamei Y., et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picard F., et al. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/S0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- 20.Cavailles V., et al. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.L’Horset F., Dauvois S., Heery D.M., Cavailles V., Parker M.G. RIP-140 interacts with multiple nuclear receptors by means of two distinct sites. Mol. Cell. Biol. 1996;16:6029–6036. doi: 10.1128/mcb.16.11.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White R., et al. The nuclear receptor co-repressor nrip1 (RIP140) is essential for female fertility. Nat. Med. 2000;6:1368–1374. doi: 10.1038/82183. [DOI] [PubMed] [Google Scholar]
- 23.Leonardsson G., et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christian M., et al. RIP140-targeted repression of gene expression in adipocytes. Mol. Cell. Biol. 2005;25:9383–9391. doi: 10.1128/MCB.25.21.9383-9391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Z.Y., et al. Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7569–7574. doi: 10.1073/pnas.1332633100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang X., Powelka A.M., Soriano N.A., Czech M.P., Guilherme A. PTEN, but not SHIP2, suppresses insulin signaling through the phosphatidylinositol 3-kinase/Akt pathway in 3T3-L1 adipocytes. J. Biol. Chem. 2005;280:22523–22529. doi: 10.1074/jbc.M501949200. [DOI] [PubMed] [Google Scholar]
- 27.Soukas A., Socci N.D., Saatkamp B.D., Novelli S., Friedman J.M. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J. Biol. Chem. 2001;276:34167–34174. doi: 10.1074/jbc.M104421200. [DOI] [PubMed] [Google Scholar]
- 28.Kelly D.P., Scarpulla R.C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 29.Weitzel J.M., Iwen K.A., Seitz H.J. Regulation of mitochondrial biogenesis by thyroid hormone. Exp. Physiol. 2003;88:121–128. doi: 10.1113/eph8802506. [DOI] [PubMed] [Google Scholar]
- 30.Lin J., et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell. 2005;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber S.N., et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mootha V.K., et al. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arner P. The adipocyte in insulin resistance: key molecules and the impact of the thiazolidinediones. Trends Endocrinol. Metab. 2003;14:137–145. doi: 10.1016/S1043-2760(03)00024-9. [DOI] [PubMed] [Google Scholar]
- 34.Nugent C., et al. Potentiation of glucose uptake in 3T3-L1 adipocytes by PPAR gamma agonists is maintained in cells expressing a PPAR gamma dominant-negative mutant: evidence for selectivity in the downstream responses to PPAR gamma activation. Mol. Endocrinol. 2001;15:1729–1738. doi: 10.1210/me.15.10.1729. [DOI] [PubMed] [Google Scholar]
- 35.Kanoh Y., Bandyopadhyay G., Sajan M.P., Standaert M.L., Farese R.V. Thiazolidinedione treatment enhances insulin effects on protein kinase C-zeta /lambda activation and glucose transport in adipocytes of nondiabetic and Goto-Kakizaki type II diabetic rats. J. Biol. Chem. 2000;275:16690–16696. doi: 10.1074/jbc.M000287200. [DOI] [PubMed] [Google Scholar]
- 36.Mukherjee R., et al. A selective peroxisome proliferator-activated receptor-gamma (PPARgamma) modulator blocks adipocyte differentiation but stimulates glucose uptake in 3T3-L1 adipocytes. Mol. Endocrinol. 2000;14:1425–1433. doi: 10.1210/me.14.9.1425. [DOI] [PubMed] [Google Scholar]
- 37.Treuter E., Albrektsen T., Johansson L., Leers J., Gustafsson J.A. A regulatory role for RIP140 in nuclear receptor activation. Mol. Endocrinol. 1998;12:864–881. doi: 10.1210/me.12.6.864. [DOI] [PubMed] [Google Scholar]
- 38.Tamori Y., Masugi J., Nishino N., Kasuga M. Role of peroxisome proliferator-activated receptor-gamma in maintenance of the characteristics of mature 3T3-L1 adipocytes. Diabetes. 2002;51:2045–2055. doi: 10.2337/diabetes.51.7.2045. [DOI] [PubMed] [Google Scholar]
- 39.Luo J., et al. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol. Cell. Biol. 2003;23:7947–7956. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higami Y., et al. Adipose tissue energy metabolism: altered gene expression profile of mice subjected to long-term caloric restriction. FASEB J. 2004;18:415–417. doi: 10.1096/fj.03-0678fje. [DOI] [PubMed] [Google Scholar]
- 41.Deveaud C., Beauvoit B., Salin B., Schaeffer J., Rigoulet M. Regional differences in oxidative capacity of rat white adipose tissue are linked to the mitochondrial content of mature adipocytes. Mol. Cell. Biochem. 2004;267:157–166. doi: 10.1023/B:MCBI.0000049374.52989.9b. [DOI] [PubMed] [Google Scholar]
- 42.Abel E.D., et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 43.Shepherd P.R., et al. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J. Biol. Chem. 1993;268:22243–22246. [PubMed] [Google Scholar]
- 44.Lowell B.B., Shulman G.I. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 45.Kelley D.E., He J., Menshikova E.V., Ritov V.B. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 46.Ojuka E.O., Jones T.E., Han D.H., Chen M., Holloszy J.O. Raising Ca2+ in L6 myotubes mimics effects of exercise on mitochondrial biogenesis in muscle. FASEB J. 2003;17:675–681. doi: 10.1096/fj.02-0951com. [DOI] [PubMed] [Google Scholar]
- 47.Boule N.G., Haddad E., Kenny G.P., Wells G.A., Sigal R.J. Effects of exercise on glycemic control and body mass in type 2 diabetes mellitus: a meta-analysis of controlled clinical trials. JAMA. 2001;286:1218–1227. doi: 10.1001/jama.286.10.1218. [DOI] [PubMed] [Google Scholar]
- 48.Petersen K.F., et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eriksson K.F., Lindgarde F. Impaired glucose tolerance in a middle-aged male urban population: a new approach for identifying high-risk cases. Diabetologia. 1990;33:526–531. doi: 10.1007/BF00404139. [DOI] [PubMed] [Google Scholar]
- 50.Wisloff U., et al. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307:418–420. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- 51.Park S.Y., et al. Depletion of mitochondrial DNA causes impaired glucose utilization and insulin resistance in L6 Glut4myc myocytes. J. Biol. Chem. 2005;280:9855–9864. doi: 10.1074/jbc.M409399200. [DOI] [PubMed] [Google Scholar]
- 52.Tiraby C., et al. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 2003;278:33370–33376. doi: 10.1074/jbc.M305235200. [DOI] [PubMed] [Google Scholar]
- 53.Cock T.A., Houten S.M., Auwerx J. Peroxisome proliferator-activated receptor-gamma: too much of a good thing causes harm. EMBO Rep. 2004;5:142–147. doi: 10.1038/sj.embor.7400082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huss J.M., Torra I.P., Staels B., Giguere V., Kelly D.P. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B.M. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 56.Meirhaeghe A., et al. Characterization of the human, mouse and rat PGC1 beta (peroxisome-proliferator-activated receptor-gamma co-activator 1 beta) gene in vitro and in vivo. Biochem. J. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diatchenko L., et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.