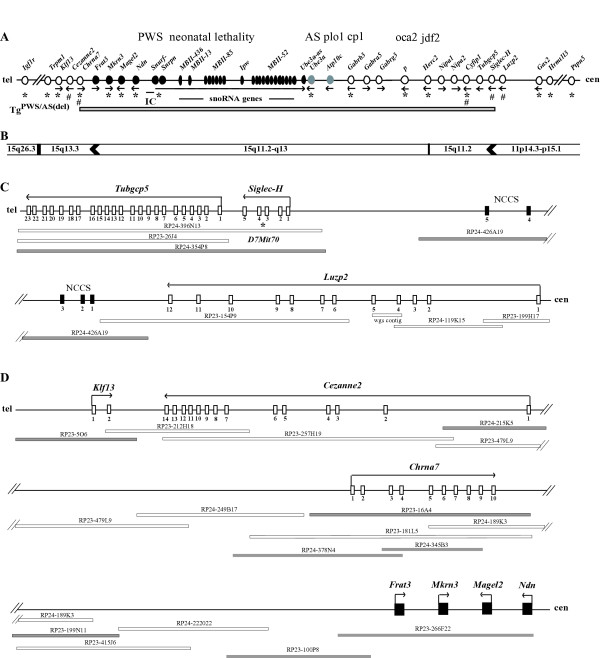
Figure 1.
Genetic and physical maps of mouse chromosome 7B/C. (A) The mouse PWS/AS-homologous region and flanking genes. Symbols are: circles, protein-coding genes; ovals, RNA-coding genes; black, paternally-expressed; grey, maternally-expressed; white, biparentally-expressed; line arrows, transcriptional orientation of genes; IC; imprinting center; plo1, p-locus-associated obesity; cp1, cleft palate 1; oca2, oculocutaneous albinism type II (p, pink-eyed dilution); jdf2, juvenile development and fertility 2; cen, centromere; tel, telomere; *, genes represented on the Gene chip MG_U74Av2 (see Table 1); #, genes analyzed by QRT-PCR; horizontal grey bar, extent of the TgPWS/TgAS ~5 Mb deletion. The first 3 genes and one more distant gene extending out from each of the centromeric and telomeric deletion breakpoints are also shown. (B) Human synteny for genes from the mouse chromosome 7B/C domain. The human chromosome locations for each orthologous mouse gene shown in (A) are given. Arrowheads represent rodent-specific gene duplications for which no ortholog is found in human. (C) BAC contig across the TgPWS/TgAS centromeric deletion breakpoint region. Symbols are: white boxes, exons; line arrows, transcriptional orientation of genes; white bars, BACs, and one "wgs" (whole genome shotgun) contig; grey bars, BACs used as FISH probes; black boxes, Non-Coding Conserved Sequences (NCCSs) in RP24-246A19; *, D7Mit70. The interval from Luzp2 through Siglec-H and Tubgcp5 is shown. (D) BAC contig across the TgPWS/TgAS telomeric deletion breakpoint region. Symbols are as for Fig 1C, except: black boxes, four intronless imprinted genes. The interval from the telomeric end of the 2 Mb imprinted gene domain through Chrna7 to Cezanne2 and Klf13 is shown.