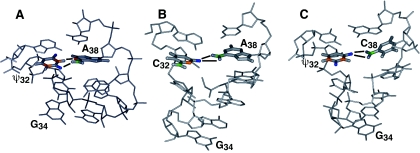
Figure 8.
Comparison of residues 31–39 of (A) the solution structure of E.coli tRNAPhe (42) and the crystal structures of (B) fully modified yeast tRNAPhe (15,16) and (C) fully modified tRNAAsp (18,21). ψ32 and A38 in (A) form a Watson–Crick base pair. In (B) and (C), nucleotides C32-A38 and ψ32-C38, respectively, form the bifurcated hydrogen bond. The anticodon loop in (A) adopts the tri-loop conformation whereas the anticodon loops in (B) and (C) adopt the U-turn motif. In E.coli tRNACys, ψ32 is in the syn configuration about the glycosidic bond and the loop forms a U-turn motif (17).