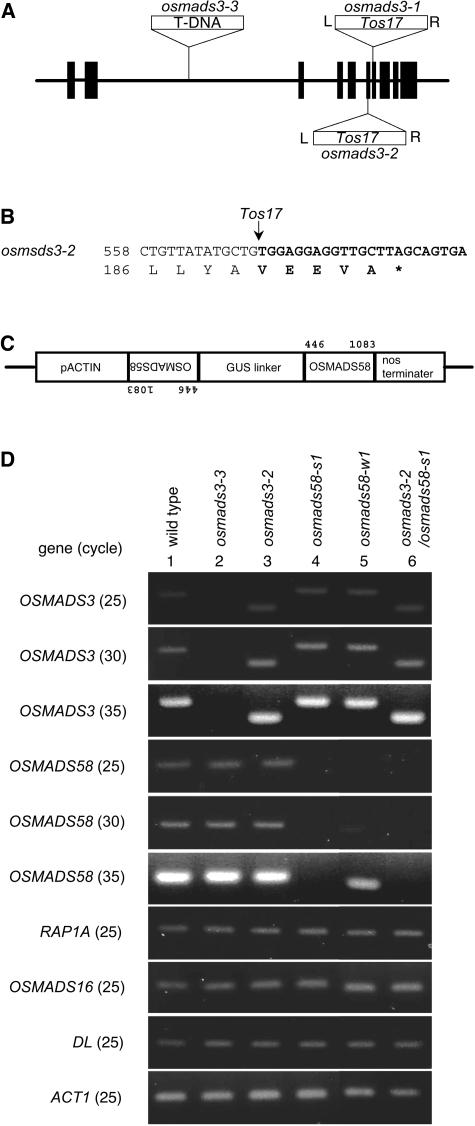
Figure 3.
Molecular Characterization of Insertional Mutants of OSMADS3 and RNAi Plants of OSMADS58.
(A) Genomic structure of OSMADS3 and position of the T-DNA or Tos17 insertion. Boxes indicate exons, and thick lines indicate introns.
(B) Insertion site of Tos17 in osmads3-2. Tos17 sequences are shown in bold. A possible chimeric translation product, terminating within the Tos17 element (asterisk), is shown beginning with amino acid 186.
(C) Schematic representation of the transgene construct, which can produce double-stranded RNA of the 3′-half the OSMADS58 sequence with a loop. The positions of the nucleotides of OSMADS58 used in this construct are shown.
(D) RT-PCR–based expression analysis of ABC MADS box genes and DL in young flowers of wild-type, osmads3 mutant, osmads58RNAi, and osmads3-2/osmads58RNAi lines. Rice ACT1 was analyzed as a control. The genes subjected to amplification are shown at the left. The number in parentheses indicates the PCR cycle number. Lines from which the RNA samples were isolated are shown at the top. To monitor OSMADS3 expression in osmads3-2, a gene-specific primer and a Tos17-specific primer were used, which resulted in a smaller fragment as compared with samples containing the wild-type allele.