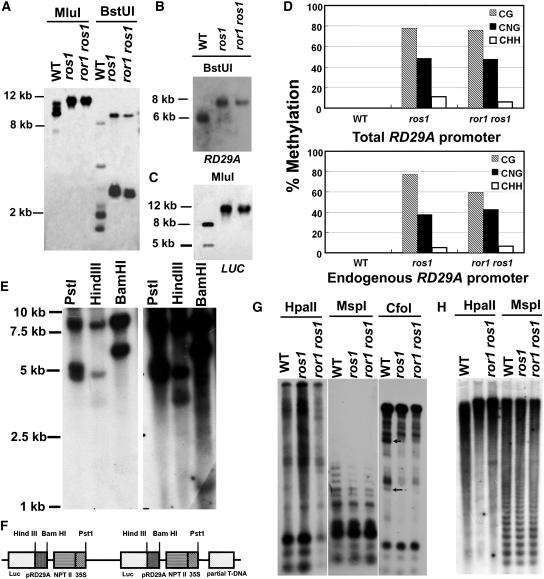
Figure 3.
ror1 Mutations Have No Effect on DNA Methylation of the RD29A Promoter, rDNA, and Centromeric DNA.
Genomic DNA samples isolated from C24 wild-type, ros1, and ror1 ros1 plants were digested with different methylation-sensitive restriction enzymes and subjected to DNA gel blot analysis or assayed by bisulfite sequencing.
(A) MluI and BstUI, with the RD29A promoter used as a probe to detect the total methylation of the RD29A promoter.
(B) BstUI, with RD29A used as a probe to detect the methylation of the endogenous RD29A promoter.
(C) MluI, with LUC used as a probe to detect the methylation of the transgene RD29A promoter.
(D) Bisulfite sequencing analysis of cytosine methylation in the upper strand of RD29A. Top: overall methylation of both the transgene RD29A promoter and the endogenous RD29A gene promoter. Bottom: methylation in endogenous RD29A promoter (Yamaguchi-Shinozaki and Shinozaki, 1994).
(E) Analysis of T-DNA copy number in ProRD29A:LUC transgenic plants. The genomic DNA was digested with restriction enzymes with unique sites in the T-DNA region, including PstI, HindIII, and BamHI, and hybridized with luciferase as probe.
(F) The deduced T-DNA insertion structure in ProRD29A:LUC transgenic plants.
(G) HpaII, MspI, and CfoI, with rDNA used as a probe. Arrows point to two extra bands in C24 wild-type plants not found in ror1 ros1 and ros1 mutant plants.
(H) HpaII and MspI, with 180-bp centromeric DNA used as a probe.