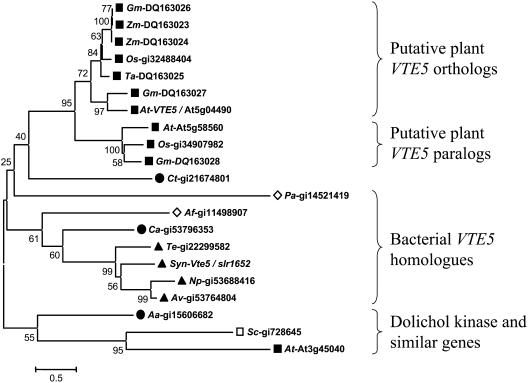
Figure 5.
Neighbor-Joining Tree of VTE5 Orthologs.
The tree was constructed from representative VTE5 orthologous sequences using a p-distance amino acid distance model and pairwise deletion. For details, see Methods. The alignment used to generate this tree is provided in Supplemental Figure 1 online. Closed squares represent plant sequences, the open square indicates yeast sequence, closed triangles represent cyanobacterial sequences, closed circles show eubacterial sequences, and open diamonds indicate archaeal sequences. The relationship of yeast (Sc) and Arabidopsis (At) dolichol kinases to the VTE5 family is also shown. The numbers on the left side indicate percent bootstrap values. The branch lengths calculated based on 5% amino acid substitutions per site are shown by the bar labeled 0.5. Reference numbers are as follows: numbers following the species abbreviations represent accession numbers; Synechocystis vte5, nucleotides 3,452,461 to 3453162, accession number gi|47118304. Aa, Aquifex aeolicus VF5; Af, Archaeoglobus fulgidus; At, Arabidopsis thaliana; Av, Anabaena variabilis; Ca, Chloroflexus aurantiacus; Ct, Chlorobium tepidum; Gm, Glycine max; Np, Nostoc punctiforme; Os, Oryza sativa; Pa, Pyrococcus abyssi; Sc, Saccharomyces cerevisiae; Syn, Synechocystis sp PCC 6803; Te, Thermosynechococcus elongatus; Zm, Zea mays.