Abstract
Autoantibody-mediated tissue destruction is among the main features of organ-specific autoimmunity. This report describes “an antibody enzyme” (abzyme) contribution to the site-specific degradation of a neural antigen. We detected proteolytic activity toward myelin basic protein (MBP) in the fraction of antibodies purified from the sera of humans with multiple sclerosis (MS) and mice with induced experimental allergic encephalomyelitis. Chromatography and zymography data demonstrated that the proteolytic activity of this preparation was exclusively associated with the antibodies. No activity was found in the IgG fraction of healthy donors. The human and murine abzymes efficiently cleaved MBP but not other protein substrates tested. The sites of MBP cleavage determined by mass spectrometry were localized within immunodominant regions of MBP. The abzymes could also cleave recombinant substrates containing encephalytogenic MBP85-101 peptide. An established MS therapeutic Copaxone appeared to be a specific abzyme inhibitor. Thus, the discovered epitope-specific antibody-mediated degradation of MBP suggests a mechanistic explanation of the slow development of neurodegeneration associated with MS.
Keywords: catalytic antibodies, Copaxone, multiple sclerosis, surface enhanced laser desorption ionization, experimental allergic encephalomyelitis
Multiple sclerosis (MS) is an autoimmune neurodegenerative disease leading to destruction of the myelin sheath that ultimately affects the ability of nerves to conduct electrical impulses (1). A poor understanding of the etiology of MS has complicated the development of effective therapeutics (2). Despite strong evidence for the contribution of T cell responses to manifestations of autoimmunity in the central nervous system (CNS) of patients with MS (3, 4), recent findings encouraged investigators to search also for B cell-mediated contributions to the MS pathogenesis (5, 6).
Ample data indicate that a significant portion of MS cases is characterized by the presence in the blood of autoantibodies against myelin protein components (7, 8). Moreover, high-resolution microscopic analysis detected myelin-specific autoantibodies in the regions of demyelination plaques in human MS and a MS-like disease of marmosets, suggesting their direct contribution to myelin destruction (9). Although the mechanism of the autoantibody role in MS pathogenesis is unknown (2), autoantibodies to myelin basic protein (MBP) and myelin oligodendrocyte glycoprotein (MOG) were proposed as biomarkers for clinical prognosis of MS (10). Similar immunoglobulins were also found in mice with induced experimental allergic encephalomyelitis (EAE), which is an animal model of MS (11). In this report, we present the evidence that anti-MBP autoantibodies of MS patients and EAE mice exhibit site-specific proteolytic cleavage of the MBP molecule that may contribute to pathological destruction of the myelin sheath.
Materials and Methods
Patients and Healthy Donors. Frozen serum samples were obtained from the Vladimirsky Moscow Region Clinical Institute. Autoantibody purification and characterization were done from the serum of 24 MS patients who had not been treated with steroids or nonsteroidal antiinflammatory drugs. The MS diagnosis was confirmed and expanded disability status scale values were calculated according to C. Poser classification (12) of disease progression by using clinical, immunological, and MRI data analysis. The sera from three patients with a high degree of the catalytic activity, and expanded disability status scale were used for kinetic and mass spectrometric analysis. Data presented for one of these patients. Blood samples of 20 healthy volunteers were used as control. An informed consent was obtained from each person as approved by the Institutional Review Board of the Vladimirsky Moscow Region Clinical Institute in accordance with the regulations of the Ministry of Health of the Russian Federation. Statistical analysis was performed according to SPSS13 software.
EAE Induction in SJL and C57BL/6 Mice. The animal work was performed in the Pushchino branch of the Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, in accordance with the regulations of the Department of Health and Human Services, National Institutes of Health Animal Welfare Insurance Reactivation no. A5230-01, duration August 14, 2000, to Aug. 31, 2005. All work was accomplished under the supervision of the Institutional Animal Care and Use Committee (U.S.) and using the Regulations of the Ministry of Health of the Russian Federation. SPF female SJL mice, 6 to 8 weeks old, were immunized according to the established protocol (13) with bovine MBP injected at 50 μg per mouse in complete Freund's adjuvant containing 2 mg/ml Mycobacterium tuberculosis. SPF female C57BL/6 mice, 6 to 8 weeks old, were immunized according to the established protocol (14) with recombinant extracellular domain of MOG injected at 100 μg per mouse in complete Freund's adjuvant containing 0.5 mg/ml M. tuberculosis. Between 14 and 28 days after a second immunization, mice with pronounced clinical symptoms were euthanized and their sera were collected for later experiments.
Antibody Purification. IgG from sera of humans and mice were purified essentially as described in refs. 15 and 16. IgGs were further separated by the antigen affinity chromatography on a column with MBP immobilized on NHS-Sepharose (Amersham Pharmacia), and their purity was assessed by electrophoresis followed by silver staining. Fab fragments were prepared as described in ref. 12 by using papain immobilized on NHS-Sepharose.
MBP Preparations. MBP was prepared from bovine brain according to ref. 12, the obtained protein was purified by reverse-phase HPLC on column C4 10/250 (Mashery-Nagel, Germany). MBP was phosphorylated by PKC as described in ref. 17 and according to the manufacturer's instructions.
Synthesis, Cloning, Expression, and Purification of ED-MOG and Trx-Fused MBP Peptides. A DNA fragment corresponding to extracellular Ig-like domain of MOG were amplified by PCR with specific primers 5′GAGGAAGCCATGGCAGGGCAGTTCAGAGTGATA3′ and 5′AGAGGAGAGATCTTCTACTTTCAATTCCATTGC3′ by using human genomic DNA as the initial template. To obtain pET32CH vector, the coding sequence of the c-myc epitope was added to pET32b+ plasmid (Novagen) by using BamHI and EcoRI restriction sites for unambiguous identification of recombinant proteins by monoclonal anti-c-myc Ab. The NcoI-BglII PCR product was cloned into pET32CH by using NcoI and BamHI restriction sites. The expression product of this plasmid that contained Trx fused to MOG1-117 (Trx-MOG) was used for cleavage analysis. The NcoI-XhoI insert of the construct was finally recloned into pET22b+ vector (Novagen) by corresponding restriction sites. Recombinant protein coded by this construct named MOG was used for immunization.
Twelve DNA fragments encoding human MBP peptides (for details, see Fig. 4) were prepared by PCR with four overlapping oligonucleotides for each other and were ligated to DNA fragment corresponding to linker (SGGGG)3S. The final PCR products were cloned in-frame into pET32CH plasmid by using NcoI and BamHI restriction sites. The expression products of these plasmids that contained Trx fused to MBP peptides were used for cleavage analysis. The plasmid encoding Trx with linker (SGGGG)3S was designed for control.
Fig. 4.
The specificity of the anti-MBP antibodies toward recombinant substrates. Schematic description of the designed fusion proteins (A) Identification of the cleavage sites of Trx-MBP fusion proteins and nonmodified Trx generated by anti-MBP antibodies: SJL mice with EAE (white boxes), MS patient (red boxes), and C57BL/6 mice with EAE (blue boxes) (B). Data obtained by mass-spectrometry analysis and gel electrophoresis of degradation products (Fig. 8, which is published as supporting information on the PNAS web site).
The soluble recombinant His-tagged proteins were obtained by Escherichia coli expression and isolated by sorption on Talon SuperFlow (BD Biosciences) column, followed by cation exchange chromatography on Mono S column (Amersham Pharmacia) at pH 5.0 and subsequent size exclusion chromatography on Superdex 75 GL 10/300 column (Amersham Pharmacia) in 150 mM NH4HCO3 buffer.
Antibody-Mediated Proteolysis. Antibodies (0.1-1 μg) were incubated at 37°C for 14 h in the final volume of 12.5 μl PBS and 0.02% NaN3 containing 1-2 μg MBP. The samples were mixed with Laemmli's buffer. The extent of MBP degradation was visualized by SDS/PAGE in Tris-glycine and Tricine buffer systems.
For the reverse-phase HPLC-MS analysis of MBP degradation products, antibodies (1 μg) were incubated at 37°C for 24 h in the final volume of 0.1 ml PBS/0.02% NaN3 containing 30 μg MBP. The reaction was stopped by adding 10% trifluoroacetic acid up to pH 2.5. A 70-μl aliquot was applied to 4.0/150 C4 column (Waters). The fractions, corresponding to major absorbance peaks, were collected and lyophilized. The samples were further redissolved in 20 μl of 50% CH3CN/0.1% trifluoroacetic acid anda7 μl aliquot of each sample was applied to H4 Protein Chip (Ciphergen). The rest was analyzed by tricine-SDS/PAGE. Surface enhanced laser desorption ionization (SELDI) analysis was performed according to the chip manufacturer's protocol.
Enzyme Kinetics Assays. MBP (11 μM) was incubated at 37°C with antibodies (67 nM) in 1.2 ml of PBS/0.02% NaN3. One hundred-microliter aliquots were taken at the indicated time, and the reaction was stopped by adding 10% trifluoroacetic acid. The samples were further chromatographed on column C4 4.0/150 (Waters). The amount of noncleaved MBP was calculated by absorbance monitoring at 280 nm. The Michaelis constant and effective catalytic constant were determined according to the Michaelis-Menten equation by using Enzyme Kinetics module 1.1 for sigmaplot software (SPSS). Discrimination of inhibition modes and calculation of inhibition constants were performed with the same software.
Alternatively, the antibodies were mixed with H-Pro-Phe-Arg-MCA or H-Ala-Ala-Phe-MCA (0.625-100 μM) in TBS/0.1% NaN3/10 mM CaCl2. Sixty-microliter samples were transferred into wells of a black PVC plate. The plate was incubated for 1-10 hat +37°C. The fluorescence intensity was measured with 2-min intervals by Tecan Genius (Tecan, Salzburg, Austria).
Size Exclusion Chromatography. Size exclusion chromatography of the purified IgG from the MS patient was performed in 6 M urea/50 mM Tris·HCl, pH 8.0 or in 100 mM Gly-HCl, pH 2.6 on Superose 6 10/300 column (Amersham Pharmacia). In case of separation in urea, the antibodies were denatured by adding solid guanidinium chloride up to 6 M immediately before applying to the column. The fractions, corresponding to the main 150-kDa peak, were renatured either by extensive dialysis (urea chromatography) or by addition of 1 M Tris (acidic chromatography).
Inhibition by Species-Specific Anti-IgG Antibodies. Purified human or murine antibodies were batch-incubated with immobilized anti-human or anti-mouse IgG (IMTEK Bio, Moscow) for 1 h at room temperature essentially as described in refs. 15 and 16. The supernatants and resin-bound fractions were further analyzed for the MBP degrading activity as described above.
Zymography. One hundred nanograms of the assayed antibody was separated on the 5-20% gradient SDS/PAGE containing 30 μg/ml of the fluorescent substrate BSA-FITC, synthesized as described in ref. 15 in the separating gel. After being washed in 2.5% Triton X-100, the gels were incubated for 72 h in 50 mM Tris·HCl (pH 7.6)/10 mM CaCl2/0.1% NaN3 solution at 37°C in the dark and visualized by ChemiImager apparatus (Alpha Innotech) by using midrange UV transillumination and a 540-nm light filter.
Immunohistochemistry. Rats, ≈1 month of age, were euthanized by CO2 inhalation. The brains were removed, embedded in the tissue-freezing medium (Leica Instruments), and frozen in isopentane cooled with liquid nitrogen to -140°C. The sections were cut at 10 μm and fixed for 10 min in 100% acetone at -20°C. The fixed sections were blocked for 1 h in PBS/0.1% Tween-20/5% FBS/2% goat serum, followed by the incubation with the purified mouse SJL and human antibody diluted 1:2 and MAB 382 (Chemicon) diluted 1:10 in PBS/0.1% Tween-20 for 1 h at room temperature. The slides were washed three times with PBS-Tween-20 and incubated with Alexa Fluor 546- or Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes) and FITC-conjugated goat anti-human antibody (IMTEK) diluted 1:500.
Western Blotting. Western blots of total rat brain tissue separated in 15% SDS/PAGE lots were incubated with human-, mouse MBP-binding antibody and MAB 382 followed by enhanced chemiluminescence (ECL) development. For the MBP degradation analysis, samples were prepared as described above and separated on 16.5% T, 0.6% C tricine-PAGE. Western blots were incubated with MAB 382 and MAB 387 in dilutions 1:2,000 and 1:500, respectively, and visualized by ECL.
MBP Structural Analysis. SELDI analysis of MBP cleavage was performed by NP20 and H4 ProteinChips and ProteinChip Reader PBS IIc (Ciphergen), according to the manufacturer's protocol. Microsequencing of MBP hydrolysis products was done by a QSTAR mass spectrometer (Ciphergen).
Inhibition Analysis. Aliquots of tested protease inhibitors were mixed with either antibody (30 nM) or bovine trypsin (0.1 nM), and MBP (4 μM) in TBS/0.1% NaN3/10 mM CaCl2. The samples were incubated for 16 h at 37°C and analyzed on 15% SDS/PAGE. The gels were stained by Coomassie and analyzed by densitometry with totallab 2.01 software (Nonlinear Dynamics, Ltd., Newcastle upon Tyne, U.K.).
Results
Anti-MBP Antibodies in the Serum of MS Patients and EAE Mice. We detected autoantibodies to MBP in the serum of 20 of 24 MS patients under study by ELISA with purified MBP (18).
We further tested whether the anti-MBP antibodies present in the sera of the MS patients and EAE mice could interact with natively folded MBP in brain tissue. Antigen affinity-purified anti-MBP antibodies from humans and mice were used for immunohistochemical analysis in frozen rat brain sections. The specific staining pattern for these two antibodies were basically identical to that seen with a reference monoclonal anti-MBP antibody. Therefore, at least some of the MBP epitopes in native myelin structures are accessible to these antibodies (Fig. 1).
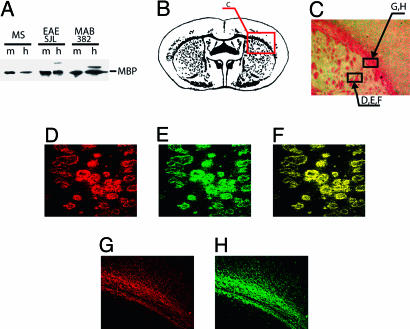
Fig. 1.
IgGs of MS patient and EAE mice react with MBP in rat brain sections. (A) Western blotting of purified MBP (m) and rat brain tissue homogenate (h) with antigen affinity-purified IgG from a MS patient, a EAE SJL mouse, and with MAB382. (B) Schematic myelin distribution in a rat brain section. (C) Wright's stain of the rat brain section chosen for analysis. D-H were taken from the indicated areas. (D-F) Double-label immunofluorescence of mouse anti-MBP monoclonal antibody MAB382 and catalytic IgG from MS patient. (D) MAB382 (Alexa 546, red). (E) MS (FITC, green). (F) Merge of D and E.(G and H) Immunofluorescence of MAB382 and IgG from EAE SJL mouse on serial sections. (G) MAB382 (Alexa 546, red). (H) IgG from EAE mouse (Alexa 488, green).
Proteolytic Activity of Purified Anti-MBP Antibodies. We noted augmented levels of the proteolytic activity in IgG fractions of the sera of MBP seropositive MS patients compared to IgGs of healthy donors (15), and, therefore, we hypothesized that autoantibodies toward MBP possessed the antigen-specific proteolytic activity. To test this hypothesis, we analyzed the proteolytic activity of the autoantibodies isolated from 24 clinically definite MS patients. Incubation of the autoantibodies with purified MBP, followed by the SDS electrophoresis analysis, revealed significant degradation of the protein. The sera from healthy donors were catalytically inactive. Importantly, the level of the proteolytic activity correlated with the expanded disability status scale range of the patients (r2 = 0.85, P < 0.001 by Spearman rank correlation) (18).
The proteolytic activity toward MBP was also detected in the antibodies isolated from the serum of SJL and C57BL/6 mice induced to develop EAE, an animal model of MS. Both human and murine autoantibodies appeared specifically to cleave MBP (Fig. 2A), but they did not exert any detectable proteolytic activity toward recombinant extracellular domain of MOG (Fig. 2B).
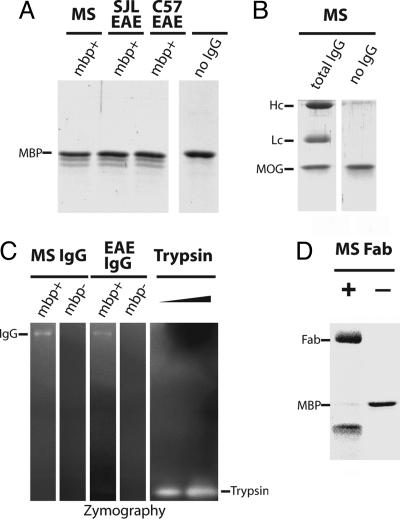
Fig. 2.
MBP degradation by purified autoantibodies. MBP affinity-purified antibodies from sera of the MS patient and EAE SJL and EAE C57BL/6 mice were incubated with purified MBP (A) and MOG (B) followed by SDS/PAGE analysis and Coomassie staining. IgG mbp+, MBP-binding antibodies. IgG heavy chains, light chains, and noncleaved MBP bands are marked as Hc, Lc, and MBP, respectively. (C) The MBP autoantibodies were separated by 5-20% gradient SDS/PAGE with FITC-BSA fluorescent substrate impregnated in the separating gel. After electrophoresis, proteins were in-gel renatured by Triton X-100 washes. Proteolytic degradation was visualized by fluorescence intensity increase, seen as bright bands on the dark background. Bovine trypsin (10 and 30 pg) was used as a positive control. IgG mbp+, MBP-binding antibodies; mbp-, antibodies not bound to the MBP-affinity column. (D) MBP degradation by the purified Fab fragment derived from the whole IgG of the MS patient visualized by SDS/PAGE.
Control experiments were performed to demonstrate that the proteolytic activity of the autoantibodies was intrinsic and did not reflect protease contamination. The MBP-specific proteolytic activity coeluted with IgG fraction isolated by size-exclusion chromatography under denaturing conditions (data not shown). When the autoantibodies were separated by SDS gel electrophoresis under nonreducing conditions and further analyzed by zymography, the proteolytic activity was detected only in the IgG band (Fig. 2C). The purified Fab fragments of the autoantibodies also retained the proteolytic activity (Fig. 2D). Finally, the proteolytic activity of the MBP-specific IgG fractions could be eliminated by adsorption on immobilized anti-human or anti-mouse Ig, respectively (data not shown). Altogether, these data demonstrated that the proteolytic activity of this preparation was associated with the antibodies and ruled out the involvement of any protease contaminant.
In the assay using highly purified bovine MBP or Pro-Phe-Arg-MCA peptide as a substrate, Michaelis-like kinetics for autoantibody-mediated proteolysis were observed. We obtained the estimates of kcat/Km = 1.1·103 M-1 sec-1 (human), 3.0·103 M-1·sec-1 (mouse) and kcat/Km = 0.75·103 M-1·sec-1 (human), 3.9·103 M-1·sec-1 (mouse) for MBP and peptide, respectively, assuming that all of the assayed IgG molecules were catalytically active (Fig. 6, which is published as supporting information on the PNAS web site). The range of catalytic efficiency of the autoantibodies was lower than in the case of sequence-specific proteases; e.g., the catalytic efficiency of bovine enterokinase was 7.1·105 M-1·sec-1 for its natural protein substrate trypsinogen (19). However, the calculated velocities of catalysis by the MBP autoantibodies are among the highest values reported for the antibody catalysis (16, 20-23) and, therefore, sufficient for their pathological effects during the slow development of neurodegeneration in the course of MS progression.
Substrate Specificity of the MBP Cleavage by Autoantibodies. To identify major sites of autoantibody-mediated proteolysis in the MBP molecule, we analyzed peptide cleavage products by a combination of reverse-phase chromatography (RP-HPLC), tricine-SDS electrophoresis, Western blot analysis, and mass-spectrometry (Fig. 3 A and B; see also Fig. 7, which is published as supporting information on the PNAS web site). The resulting data revealed the presence of six sites of preferential proteolysis; all of them were located after Arg or Lys (Fig. 3C). Cleavage at these sites occurred at a similar rate as determined by 32P-MBP degradation analysis (Fig. 3D). Interestingly, most of the sites (5 of 6) of the cleavage were located within the immunodominant regions of MBP (Fig. 3C, marked by yellow (24); two of them falling inside the sequence fragment spanning from the 86 to 98 residues (Fig. 3C, marked by the red box) that proved to be a specific inducer of EAE in SJL mice (25).
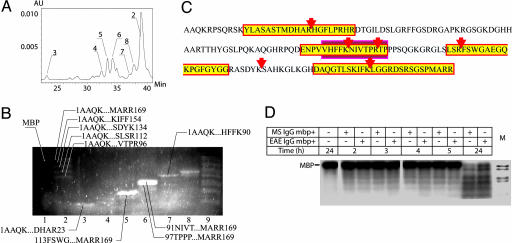
Fig. 3.
Analysis of major MBP cleavage products. Reverse-phase HPLC-MS analysis of major MBP cleavage products. Column eluate fractions, corresponding to dominant chromatography peaks (A), were collected, freeze dried, redissolved, and applied to SELDI H4 chip and tricine-SDS/PAGE (B). Gel was stained by Sypro Orange. Peptides, unambiguously identified by SELDI and clearly seen in corresponding gel lanes, are indicated. (C) Schematic description of the preferential antibody cleavage sites in the MBP sequence. Sequence fragments identical to the immunodominant MBP-derived peptides (12-31, 82-98, 110-128, and 144-169) are shown in yellow rectangles, and the encephalitogenic peptide region (86-98) is marked by the red box. (D) 32P-MBP degradation by autoantibodies. Autoradiography of 32P-phosphorylated MBP hydrolysis by proteolytic mouse (EAE SJL) and human (MS) IgG. Line M, molecular mass markers (range 2.5-16.9 kDa, Amersham Pharmacia).
To test the abzyme specificity toward distinct MBP fragments, 12 recombinant fusion proteins of Trx with the C-terminally fused MBP peptides were designed (Fig. 4). The products of hydrolysis were analyzed by SDS electrophoresis (Fig. 8) and by mass spectrometry. Only two recombinant substrates (Fig. 4B, 7 and 8) containing encephalitogenic MBP85-101 peptide were readily hydrolyzed by the anti-MBP autoantibodies from MS patients and C57BL/6 mice with induced EAE. Fusion proteins (1 and 3-12) were cleaved by autoantibodies isolated from SJL mice with induced EAE. In all cases, recombinant substrates were cleaved only at preferential sites inside the MBP fragment, leaving the Trx part undegraded (Fig. 8). In contrast, limited proteolysis of these substrates with enterokinase and trypsin (Fig. 8) resulted in a cleavage pattern with multiple products, indicating an efficient Trx cleavage. These data further confirmed the substrate specificity of the antibody and its profound difference from trypsin, a common protease that cleaves at basic residues.
Inhibition Analysis of Antibody-Mediated MBP Cleavage. Recently, the antibody-antigen interaction was shown to trigger the peroxidation pathway (23) that might mediate oxidative antigen degradation. To test the possibility of autoantibody-mediated MBP proteolysis via the peroxidation pathway, we performed the cleavage reactions under nonoxigenic conditions by using inhibitors to block H2O2 generation by IgG.
At the IC50 inhibitor concentration (26) and even 10 times higher, they displayed no significant effect on autoantibody-mediated MBP and fused peptide proteolysis (Fig. 9, which is published as supporting information on the PNAS web site). Similarly, no effect was observed under oxygen-free conditions (26). Thus, the cleavage of MBP by the autoantibodies is not caused by the autoantibody-mediated peroxide formation.
The chemical mechanism of the antibody proteolytic activity was further probed with a set of known protease inhibitors. Degradation of MBP (Table 1) and Pro-Phe-Arg-MCA peptide was irreversibly inhibited by the Ser- and His-reactive covalent inhibitors 4-(2-aminoethyl)-benzenesulfonyl fluoride (AEBSF), N-α-tosyl-l-phenylalanine chloromethyl ketone (TPCK), and N-α-tosyl-l-lysine chloromethyl ketone (TLCK), whereas reversible inhibitors of serine proteases, leupeptin, antipain, and chymostatin, were relatively ineffective. Interestingly, the catalytic activity was not affected by equimolar concentrations of aprotinin, a tight-binding inhibitor of trypsin-like proteases. It was not inhibited at all by bestatin, E-64, N-ethylmaleimide, pepstatin, and EDTA, specific inhibitors of aminopeptidases, cysteine, aspartic, and metalloproteases, respectively. We may conclude that the comparable inhibition rates of the autoantibody and trypsin activity (Table 2, which is published as supporting information on the PNAS web site) with AEBSF, TLCK, and TPCK are indicative of a similar general structure of their active sites, in particular, the presence of a nucleophilic Ser-His pair. At the same time, the observed differences in the inhibition patterns for the autoantibody and trypsin may reflect the lack of the evolutionarily related similarities between the surface shapes of their active sites.
Table 1. Inhibition of MBP-cleaving activity.
| Mode of inhibition | Proteases classes affected | Inhibitors | MS* | EAE SJL* | Bovine trypsin* |
|---|---|---|---|---|---|
| Covalent | Ser-, Cys- | AEBSF | 3·10-5 | 1.3·10-4 | 2·10-5 |
| TPCK | 1·10-3 | 1·10-3 | 1·10-3 | ||
| TLCK | 3·10-3 | 7·10-3 | 1·10-4 | ||
| Leu-CK | Inactive | Inactive | Inactive | ||
| Tight binding, covalent | Broad spectrum | α2-macroglobulin | n/d† | 3·10-9 | <2·10-10 |
| Tight binding | Trypsin-like | Aprotinine | Inactive | Inactive | <5·10-10 |
| Reversible | Trypsin-like, Cys- | Leupeptin | 2·10-5 | 3·10-4 | 4·10-6 |
| Ser-, Cys- | Antipain | 1·10-4 | 1·10-4 | <3·10-6 | |
| Chymotrypsin-like | Chymostatin | 6·10-4 | 5·10-5 | n/d | |
| Aminopeptidases | Bestatin | Inactive | Inactive | n/d | |
| Aspartic | Pepstatin | Inactive | Inactive | n/d | |
| Cys- | E-64 | Inactive | Inactive | n/d | |
| Bivalent metal ion chelator | Metalloproteases | EDTA | Inactive | Inactive | n/a |
| Modifier of free Cys residues | Cys- | N-ethylmaleimide | Inactive‡ | Inactive‡ | n/d |
Maximum concentration of inhibitors used as follows: aprotinine, 0.15 μM; leucine chlormethylketone, 1 mM; E-64, 400 μM; α2-macroglobulin, 0.6 μM (as monomer); pepstatine, 10 μM; N-ethylmaleimide, 1 μM; EDTA, 1 mM. AEBSF, 4-(2-aminoethyl)-benzenesulfonyl fluoride; TPCK, N-α-tosyl-l-phenylalanine chloromethyl ketone and TLCK, N-α-tosyl-lysine chloromethyl ketone. n/d, not determined; n/a, not applicable due to calcium stabilization of the enzyme's active form.
IC50 inhibitors concentration was determined by utilizing MBP as a substrate and given in M
IC50 is ≈ 10–30 nM by data extrapolation; protease contamination of commercially available α2-macroglobulin prevents exact determination
Without affecting catalysis, concentration dependence was not expected and observed
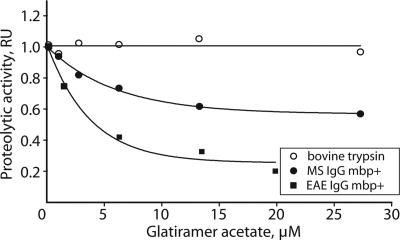
We also tested whether the autoantibody catalytic activity could be affected by glatiramer acetate (Copaxone). This substance is an effective and a specific anti-MS drug reducing the relapse rate in MS patients (27). Although the precise mechanism of this drug still remains unknown, it appears to act against the immunodominant epitope of MBP by T cell receptor antagonism and by blocking the corresponding MHC complex (28). Our tests revealed that Copaxone effectively inhibited the MBP degradation catalyzed either by human or murine IgG, but it had no effect on the trypsin cleavage of MBP (Fig. 5). By the ELISA assay, we also showed that Copaxone effectively competed with catalytic IgG for MBP binding, indicating the mechanism of its inhibitory effect. Thus, the autoantibody inhibition with Copaxone establishes a potential link of anti-MBP autoantibodies with the etiology of MS.
Fig. 5.
Proteolytic activity of the MS patient and EAE mice antibodies is inhibited by glatiramer acetate (Copaxone). Purified MBP was incubated with either trypsin or IgGs of human (MS) or mouse (EAE) and a variable concentration of glatiramer acetate (Copaxone). The proteolytic activity was calculated as the ratio of degraded and nondegraded MBP measured by the densitometry of Coomassie-stained gel.
Discussion
Autoantibody mediated tissue destruction is among the main features of organ-specific autoimmunity (29). Since the original discovery of catalytic antibodies (30-32), ample data established their contribution to pathological effects in disease (33) as well as their possible biomedical applications (23, 34). It was shown that elevated levels of the autoantibody catalytic activity correlated with an autoimmune response in human and mice models (16, 21, 35). Moreover, data from several laboratories indicate that autoantibodies can readily penetrate the blood-brain barrier (36-39).
We detected the intrinsic proteolytic activity of anti-MBP autoantibodies purified from sera of MS patients. IgGs of healthy donors did not show such an activity. Multiple control experiments established that the proteolytic activity was a property of the autoantibody rather than an indicator of the contaminant protease presence. Although the antibody proteolytic activity was significantly lower than that of common proteases, it was fairly high as compared to other catalytic antibodies. The activity was quite specific toward MBP; several other proteins were resistant to cleavage. Analysis of the sites of MBP cleavage suggested that the antibody preferentially cleaved at basic residues. However, the cleavage efficiency was also defined by the surrounding sequence and probably reflected the necessity of initial antibody recognition of a particular epitope. The epitope specificity of isolated abzymes was shown by using a set of designed recombinant substrates. Antibodies isolated from MS patients and C57BL/6 mice with EAE induced by MOG revealed strong cleavage specificity inside encephalitogenic MBP85-101 peptide. Additional cleavage sites were revealed by antibodies from SJL mice with MBP-induced EAE. This observation may be attributed to the induction of the broad catalytic antibody repertoire in the latter example. In contrast, proteolysis with trypsin did not show any preferential specificity to MBP.
Our finding that anti-MBP autoantibodies possess the intrinsic proteolytic activity specifically directed toward MBP suggests that the anti-MBP autoantibodies can recognize and destroy myelin structures by MBP cleavage. This concept would provide a mechanistic explanation of their role in pathological destruction of the myelin sheath observed in MS. The postulated importance of the autoantibody catalytic property is supported by observation of a similar activity detected in the antibodies of mice with EAE, an established animal model of MS.
We have also noted a correlation between the autoantibody catalytic activity and expanded disability status scale parameter (18). Thus, the level of catalytic antibodies in serum may provide a clinically important marker for disease progression. The inhibition of the anti-MBP catalytic antibody with Copaxone also correlates with the proposed role of their proteolytic activity in the slow progression of the disease. A search for other effective inhibitors of anti-MBP catalytic antibodies therefore may provide a previously uncharacterized approach for developing an effective treatment of MS.
Supplementary Material
Acknowledgments
We thank Ciphergen, Inc. (The ProteinChip Company) for valuable help in SELDI analysis, Teva for Copaxone samples, and Prof. Vladimir Skulachev for valuable discussion on the initial stage of this project. This work was supported, in part, by Biotechnology Engagement Program Grant 37 (to H.C.M. and A.G.G.), Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (to H.C.M. and A.G.G.), European Union International Cooperation-COPERNICUS Program Grant IC15 CT96-0909 (to A.F. and A.G.G.), International Center for Genetic Engineering and Biotechnology Grant CRP/RUS04-03 (to A.G.G.), Scientific Russian Schools Program Grant 1800.2003.4 (to A.G.G.), Program on Fundamental Medicine of the Russian Academy of Sciences Grant (to A.G.G.), and Russian Foundation of Basic Research Grant 03-04-48836-a (to N.A.P.).
Author contributions: N.A.P. and A.G.G. designed research; O.M.D., I.I.V., A.A.B., I.N.K., M.A.L., V.M.G., M.V.S., P.T., and A.K. performed research; A.A.B., G.B.T., and M.A.L. contributed new reagents/analytic tools; N.A.P., I.I.V., A.A.B., A.G.P., S.V.S., S.L.K., V.M.G., B.A., A.K., H.C.M., D.T., and A.F. analyzed data; and A.G.P., H.C.M., and A.G.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: MBP, myelin basic protein; MOG, myelin oligodendrocyte glycoprotein; MS, multiple sclerosis; SELDI, surface enhanced laser desorption ionization.
References
- 1.Schwartz, R. S. (1993) in Fundamental Immunology, ed. Paul, W. E. (Raven, New York), pp. 1033-1097.
- 2.Hafler, D. A. (2004) J. Clin. Invest. 113, 788-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofstetter, H. H., Sewell, D. L., Liu, F., Sandor, M., Forsthuber, T., Lehmann, P. V. & Fabry, Z. (2003) J. Neuroimmunol. 134, 25-34. [DOI] [PubMed] [Google Scholar]
- 4.Hohlfeld, R. & Wekerle, H. (2001) Curr. Opin. Neurol. 14, 299-304. [DOI] [PubMed] [Google Scholar]
- 5.Archelos, J. J., Storch, M. K. & Hartung, H. P. (2000) Ann. Neurol. 47, 694-706. [PubMed] [Google Scholar]
- 6.Correale, J. & de los Milagros Bassani Molinas, M. (2002) J. Neurol. 249, 375-389. [DOI] [PubMed] [Google Scholar]
- 7.Chamczuk, A. J., Ursell, M., O'Connor, P., Jackowski, G. & Moscarello, M. A. (2002) J. Immunol. Methods 262, 21-27. [DOI] [PubMed] [Google Scholar]
- 8.Reindl, M., Linington, C., Brehm, U., Egg, R., Dilitz, E., Deisenhammer, F., Poewe, W. & Berger, T. (1999) Brain 122, 2047-2056. [DOI] [PubMed] [Google Scholar]
- 9.Genain, C. P., Cannella, B., Hauser, S. L. & Raine, C. S. (1999) Nat. Med. 5, 170-175. [DOI] [PubMed] [Google Scholar]
- 10.Berger, T., Rubner, P., Schautzer, F., Egg, R., Ulmer, H., Mayringer, I., Dilitz, E., Deisenhammer, F. & Reindl, M. (2003) N. Engl. J. Med. 349, 139-145. [DOI] [PubMed] [Google Scholar]
- 11.Fritz, R. B., Chou, C. H. & McFarlin, D. E. (1983) J. Immunol. 130, 191-194. [PubMed] [Google Scholar]
- 12.Poser, C. M. & Brinar, V. V. (2001) Clin. Neurol. Neurosurg. 103, 1-11. [DOI] [PubMed] [Google Scholar]
- 13.Miller, S. D. & Karpus, J. W. (1996) in Current Protocols in Immunology, ed. Coligan, J. E. (Wiley, New York), Vol. 3, pp. Unit 15.1.1-15.1.13. [Google Scholar]
- 14.Oliver, A. R., Lyon, G. M. & Ruddle, N. H. (2003) J. Immunol. 171, 462-468. [DOI] [PubMed] [Google Scholar]
- 15.Ponomarenko, N. A., Durova, O. M., Vorobiev, I. I., Aleksandrova, E. S., Telegin, G. B., Chamborant, O. G., Sidorik, L. L., Suchkov, S. V., Alekberova, Z. S., Gnuchev, N. V. & Gabibov, A. G. (2002) J. Immunol. Methods 269, 197-211. [DOI] [PubMed] [Google Scholar]
- 16.Shuster, A. M., Gololobov, G. V., Kvashuk, O. A., Bogomolova, A. E., Smirnov, I. V. & Gabibov, A. G. (1992) Science 256, 665-667. [DOI] [PubMed] [Google Scholar]
- 17.Kishimoto, A., Nishiyama, K., Nakanishi, H., Uratsuji, Y., Nomura, H., Takeyama, Y. & Nishizuka, Y. (1985) J. Biol. Chem. 260, 12492-12499. [PubMed] [Google Scholar]
- 18.Ponomarenko, N. A., Durova, O. M., Vorobiev, I. I., Belogurov, A. A., Telegin, G. B., Suchkov, S. V., Misikov, V. K., Morse, H. C., III, & Gabibov, A. G. (November 4, 2005) Immunol. Lett., 10.1016/j.imlet.2005.10.006. [DOI] [PubMed]
- 19.Lu, D., Yuan, X., Zheng, X. & Sadler, J. E. (1997) J. Biol. Chem. 272, 31293-31300. [DOI] [PubMed] [Google Scholar]
- 20.Gololobov, G. V., Chernova, E. A., Schourov, D. V., Smirnov, I. V., Kudelina, I. A. & Gabibov, A. G. (1995) Proc. Natl. Acad. Sci. USA 92, 254-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul, S., Volle, D. J., Beach, C. M., Johnson, D. R., Powell, M. J. & Massey, R. J. (1989) Science 244, 1158-1162. [DOI] [PubMed] [Google Scholar]
- 22.Pillet, D., Paon, M., Vorobiev, I. I., Gabibov, A. G., Thomas, D. & Friboulet, A. (2002) J. Immunol. Methods 269, 5-12. [DOI] [PubMed] [Google Scholar]
- 23.Wentworth, P., Jr., McDunn, J. E., Wentworth, A. D., Takeuchi, C., Nieva, J., Jones, T., Bautista, C., Ruedi, J. M., Gutierrez, A., Janda, K. D., et al. (2002) Science 298, 2195-2199. [DOI] [PubMed] [Google Scholar]
- 24.Sospedra, M. & Martin, R. (2005) Annu. Rev. Immunol. 23, 683-747. [DOI] [PubMed] [Google Scholar]
- 25.Sakai, K., Zamvil, S. S., Mitchell, D. J., Lim, M., Rothbard, J. B. & Steinman, L. (1988) J. Neuroimmunol. 19, 21-32. [DOI] [PubMed] [Google Scholar]
- 26.Oyanagui, Y., Taniguchi, M., Iwata, M. & Murakami, M. (2003) Life Sci. 73, 1333-1346. [DOI] [PubMed] [Google Scholar]
- 27.Arnon, R. & Sela, M. (2003) J. Mol. Recognit. 16, 412-421. [DOI] [PubMed] [Google Scholar]
- 28.Aharoni, R., Teitelbaum, D., Arnon, R. & Sela, M. (1999) Proc. Natl. Acad. Sci. USA 96, 634-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viau, M. & Zouali, M. (2005) Clin. Immunol. 114, 17-26. [DOI] [PubMed] [Google Scholar]
- 30.Lerner, R. A., Benkovic, S. J. & Schultz, P. G. (1991) Science 252, 659-667. [DOI] [PubMed] [Google Scholar]
- 31.Raso, V. & Stollar, B. D. (1975) Biochemistry 14, 591-599. [DOI] [PubMed] [Google Scholar]
- 32.Tramontano, A., Janda, K. D. & Lerner, R. A. (1986) Science 234, 1566-1570. [DOI] [PubMed] [Google Scholar]
- 33.Lacroix-Desmazes, S., Bayry, J., Misra, N., Horn, M. P., Villard, S., Pashov, A., Stieltjes, N., d'Oiron, R., Saint-Remy, J. M., Hoebeke, J., et al. (2002) N. Engl. J. Med. 346, 662-667. [DOI] [PubMed] [Google Scholar]
- 34.Mets, B., Winger, G., Cabrera, C., Seo, S., Jamdar, S., Yang, G., Zhao, K., Briscoe, R. J., Almonte, R., Woods, J. H. & Landry, D. W. (1998) Proc. Natl. Acad. Sci. USA 95, 10176-10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tawfik, D. S., Chap, R., Green, B. S., Sela, M. & Eshhar, Z. (1995) Proc. Natl. Acad. Sci. USA 92, 2145-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friden, P. M., Walus, L. R., Watson, P., Doctrow, S. R., Kozarich, J. W., Backman, C., Bergman, H., Hoffer, B., Bloom, F. & Granholm, A. C. (1993) Science 259, 373-377. [DOI] [PubMed] [Google Scholar]
- 37.Merkler, D., Oertle, T., Buss, A., Pinschewer, D. D., Schnell, L., Bareyre, F. M., Kerschensteiner, M., Buddeberg, B. S. & Schwab, M. E. (2003) FASEB J. 17, 2275-2277. [DOI] [PubMed] [Google Scholar]
- 38.Tsukada, N., Tanaka, Y., Miyagi, K., Yanagisawa, N. & Okano, A. (1989) J. Neuroimmunol. 24, 41-46. [DOI] [PubMed] [Google Scholar]
- 39.Yednock, T. A., Cannon, C., Fritz, L. C., Sanchez-Madrid, F., Steinman, L. & Karin, N. (1992) Nature 356, 63-66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.