Abstract
Many known antibiotics target the translational apparatus, but none of them can selectively inhibit initiation of protein synthesis and/or is prokaryotic-specific. This article describes the properties of GE81112, an effective and prokaryotic-specific initiation inhibitor. GE81112 is a natural tetrapeptide produced by a Streptomyces sp. identified by an in vitro high-throughput screening test developed to find inhibitors of the prokaryotic translational apparatus preferentially acting on steps other than elongation. In vivo GE81112 inhibits protein synthesis but not other cell functions such as DNA duplication, transcription, and cell wall synthesis. In vitro GE81112 was found to target the 30S ribosomal subunit and to interfere with both coded and noncoded P-site binding of fMet-tRNA, thereby selectively inhibiting formation of the 30S initiation complex.
Keywords: 30S initiation complex, fMet-tRNA binding, P-site inhibition
Despite the fact that more than half of known antibiotics target the protein synthetic machinery of the bacterial cell (1–4), the translational apparatus still offers remarkable opportunities for identifying unique inhibitors capable of bypassing existing resistance mechanisms. This possibility is due not only to the unmatched structural and functional complexity of this machinery but also to the fact that many of its components and several individual steps of the translational pathway represent unexploited antibiotic targets (3, 4).
In the present report, we describe the properties of an antibiotic designated GE81112. This antibiotic was found by a high-throughput screening procedure devised to identify preferentially, among the secondary metabolites produced by a microbial library, inhibitors targeting translational steps other than elongation. The data of this article will show that GE81112 indeed represents a structurally unique tetrapeptide antibiotic, capable of inhibiting selectively the P-site binding of fMet-tRNA, thereby blocking the formation of the 30S initiation complex. In light of its properties, GE81112 can be regarded as the most efficient, selective, and specific inhibitor of bacterial translation initiation, by far superior to all translation initiation inhibitors known so far.
Materials and Methods
Buffers. The following buffers were used: buffer A [20 mM Tris·HCl, pH 7.7/7 mM Mg(OAc)2/80 mM NH4Cl/0.1 mM 1,4-DTT], buffer B [20 mM Hepes, pH 7.1/7 mM Mg(OAc)2/80 mM NH4Cl/0.1 mM 1,4-DTT], buffer C [10 mM Tris·HCl, pH 7.7/15 mM NH4Cl/180 mM KCl/0.1 mM 1,4-DTT], and buffer D [20 mM Tris·HCl, pH 7.7/10 mM Mg(OAc)2/80 mM NH4Cl/0.1 mM 1,4-DTT].
Assessment of the in Vivo Activity of GE81112. Escherichia coli SS5012 (opp–) cells were grown at 37°C in minimal medium to OD600 = 0.2, at which point (time = 0) the culture was divided into four identical aliquots and each of them received one of the following precursors: [3H]thymidine, [3H]uridine, [14C]phenylalanine, or N-acetyl[3H]glucosamine. After 10 min, each radiolabeled culture was split into two aliquots, one of which was exposed to GE81112 (1.3 mg/ml) and the other to DMSO (final concentration 1%). At 10-min intervals, 50-μl samples of each culture were withdrawn and mixed with 50 μl of 2% sodium dodecylsulfate, and the hot acid (trichloroacetic acid)-insoluble radioactivity present in 50 μl of the resulting cell extracts was determined by liquid scintillation counting.
fMet-tRNA Binding to 30S Ribosomal Subunit. Reaction mixtures (30 μl of buffer A) contained 0.5 mM GTP, and 30 pmol of 30S ribosomal subunits. After a brief incubation in the presence of the inhibitors (as indicated in the appropriate figures), 30 pmol each of initiation factor 1 (IF1), IF2, and IF3 were added, and binding was triggered by addition of 45 pmol each of 027mRNA (a model mRNA having the same 5′ UTR as 022mRNA and the coding sequence of 002mRNA amplified 3.6 times) and f[35S]Met-tRNA. After 10-min incubation at 37°C, 30S-bound f[35S]Met-tRNA present in 20 μl of reaction mixture was determined by nitrocellulose filtration.
Kinetics of fMet-tRNA Binding to 30S Ribosomal Subunit. The analysis was carried out by using a Biologic (SFM-400; Grenoble, France) quench-flow apparatus operating in a rapid filtration mode. Syringe A contained, in 2 ml of buffer B, 0.5 mM GTP, 1 μM 30S subunits, 1.5 μM each of IF1, IF2, and IF3, and the indicated amounts of GE81112. Syringe B contained, in 2 ml of buffer B, 0.5 mM GTP, 2 μM 022mRNA (5), and 2 μM f[35S]Met-tRNA. Equal volumes (50 μl) of the two solutions were shot into the mixing chamber and incubated at 20°C for times between 30 and 5,000 ms before being rapidly filtered through a nitrocellulose disk. The amount of 30S-bound f[35S]Met-tRNA was determined by liquid scintillation counting.
Primary in Vitro Translation Tests. These tests were carried out for 60 min at 37°C on 96-well microtiter plates in reaction mixtures (50 μl) containing (i) bacterial system: 12 mM Mg(OAc)2, 100 mM NH4Cl, 10 mM Tris·HCl (pH 7.7), 2 mM DTT, 2 mM ATP, 0.4 mM GTP, 10 mM phosphoenolpyruvate, 25 μg/ml pyruvate kinase, 0.12 mM citrovorum factor (Serva), and 150 μg of total E. coli MRE600 tRNA and E. coli MRE600 S30 extract (5–7); or (ii) yeast system: 33 mM Hepes–KOH (pH 7.4), 160 mM KOAc, 3.8 mM Mg(OAc)2, 3.3 mM DTT, 0.5 mM ATP, 0.1 mM GTP, 30 mM phosphocreatine (Sigma), 20 μg/ml creatine kinase (Sigma), 200 units/ml RNase inhibitor (Promega), and 0.8–1.3 A260 units of Saccharomyces cerevisiae S30 extract. In addition, the reaction mixtures for both bacterial and yeast translation contained 200 μM each amino acid (except phenylalanine), 45 μM [14C]phenylalanine (93.8 mCi/mmol; 1 Ci = 37 GBq) or 0.5 μCi of [3H]phenylalanine (2.5 mCi/μmol) and were programmed with either 20 μg of poly(uridylic acid) [poly(U)] or an optimized amount (≈20 pmol) of 027mRNA, a derivative of 022mRNA (5), whose sequence will be reported elsewhere. After 1-h incubation at 37°C, the hot trichloroacetic acid-insoluble radioactivity was determined as described above. The reactions were stopped by 30-min incubation at 20°C after addition of 25 μl of 3 M NaOH. The trichloroacetic acid (10%)-insoluble radioactivity present in each well was recovered on a glass fiber filter (Unifilter-96, GF/B, Packard) by using a microplate harvester (Filtermate 196, Packard) and determined with a TopCount Filtration unit (Packard) after addition of a scintillation mixture (Packard; Microscint 20).
Luciferase Synthesis. Each reaction mixture (50 μl of buffer C) contained 0.4 mM GTP, 2 mM ATP, 10 mM phosphoenolpyruvate, 0.025 μg/μl pyruvate kinase, 2 μM coelenterazine, 0.2 mM each amino acid, 0.12 mM citrovorum factor, 1 μg/μl E. coli MRE600 total tRNA (Sigma), 15 pmol of Renilla luciferase mRNA, 12 μlof S30 cell extract (corresponding to ≈15 pmol of 70S ribosomes), and GE81112 as indicated. The luminescence in each luciferase reaction was recorded at 2-min intervals by using a 1450 Microbeta (Wallac, Gaithersburg, MD).
Chemical Probing and Primer Extension Analysis. Chemical reactions with dimethyl sulfate (Kodak) and kethoxal (Research Organics), extraction, and primer extension analysis of 16S rRNA were carried out as described in ref. 8.
Results
A library of Actinomycetes secondary metabolites was screened by a high-throughput test designed to identify antibacterial agents active in a bacterial cell-free system, preferentially on steps of translation other than elongation. The effect of each library sample was tested in a system programmed with a model mRNA (027mRNA) having all of the relevant characteristics of a natural template and whose expression requires a functional translation initiation mechanism. The results of this test were then compared with those obtained in the traditional poly(U)-encoded translation system, which depends only on the elongation function and on the aminoacylation of a single tRNA.
Primary screening of ≈25,000 samples yielded a fairly large number of hits. However, the majority of these were discarded because they (i) inhibited prokaryotic translation with less than two orders of magnitude preference with respect to that of yeast; (ii) inhibited poly(U)- and 027mRNA-dependent translation to a similar extent; (iii) were found to correspond to already known antibiotics; or (iv) did not yield consistent results upon refermentation of the producing strains and retesting of their activity. Thus, only two compounds were found to have the desired characteristics and eventually proved to be novel. The stronger inhibition of 027mRNA-dependent translation compared with poly(U)-dependent polyphenylalanine synthesis suggested that, rather than elongation, these compounds interfere with translation initiation and/or termination. Because the latter steps represent largely underexploited antibiotic targets, both hits were further investigated. The functional characteristics of the antibiotic designated GE81112 is reported in this article, whereas the properties of the second molecule, designated GE82832, will be reported elsewhere (L.B., A.F., M. Di Stefano, A. Lazzarini, M.A., and C. O. Gualerzi, unpublished data).
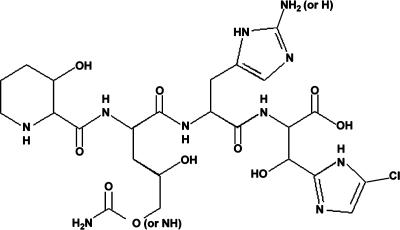
Structural analysis carried out on GE81112 primarily by NMR spectroscopy (A. Lazzarini, L.B., L. Cavalletti, M.A., E. Corti, I. Ciciliato, L. Gastaldo, A., Marazzi, M. Feroggio, A. Maio, et al., unpublished data) demonstrated that this antibiotic is a complex of three major factors (A, B, and B1), with molecular masses of 643–658 Da, and consists of a noncyclic chlorine-containing tetrapeptide whose structure (Fig. 1) does not resemble any known antibiotic. GE81112 is constituted by a histidine and three uncommon amino acids (i.e., 3-hydroxypipecolic acid, 2-amino-5-[(aminocarbonyl)oxy]-4-hydroxypentanoic acid, and 5-chloro-2-imidazolylserine) and displays a marked hydrophilic character. The results presented in this article were obtained with factor B (658 Da), which is the most active form of GE81112.
Fig. 1.
Structure of GE81112. The structure, determined by NMR spectroscopy (A. Lazzarini, L.B., L. Cavalletti, M. Abbondi, E. Corti, I. Ciciliato, L. Gastaldo, A. Marazzi, M. Feroggio, A. Maio, et al., unpublished data), consists of four amino acids (3-hydroxypipecolic acid, 2-amino-5-[(aminocarbonyl)oxy]-4-hydroxypentanoic acid, histidine, and 5-chloro-2-imidazolylserine). Three forms of GE81112 having molecular masses between 643 and 658 Da have been identified; factor B is shown in the figure with the alternative substituents of the other factors (indicated in the parentheses) present in the corresponding positions of the molecule.
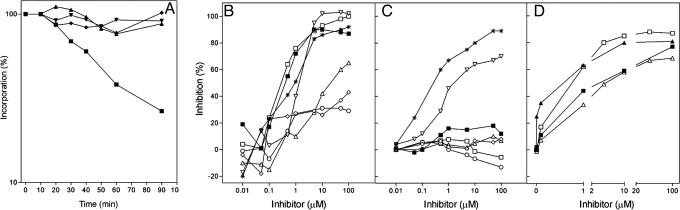
To determine whether GE81112 is capable of inhibiting protein synthesis also in vivo and whether it might also affect other cellular activities, its effect was tested on DNA, RNA, protein, and cell wall biosynthesis. Thus, four E. coli cultures each growing in the presence of a different radioactive precursor were divided into two portions upon reaching the early exponential growth phase, one serving as a control and the other receiving an amount of GE81112 corresponding to >102 times the minimum inhibitory concentration. The incorporation of the precursors into their respective products in the presence and absence of antibiotic was then followed as a function of time. As seen in Fig. 2A, GE81112 had no effect on DNA duplication, transcription, and cell wall biosynthesis but caused instead an immediate blockage of protein synthesis. This result indicates that GE81112 is a selective and powerful translation inhibitor also in vivo.
Fig. 2.
Inhibitory effects of GE81112 in vivo and in vitro. (A) Effect of GE81112 on the in vivo incorporation of [3H]thymidine (▾), [3H]uridine (▴), [14C]phenylalanine (▪), and N-[3H]acetylglucosamine (♦) by E. coli cells. (B) Comparison of the effect of GE81112 and of other antibiotics on 027mRNA-directed E. coli translational systems in vitro: GE81112 (▪), viomycin (□), edeine (▿), pactamycin ( ), linezolid (⋄), sisomicin (○), and kasugamycin (▵). (C) Comparison of the effect of GE81112 and of other antibiotics on 027mRNA-directed S. cerevisiae translational systems in vitro: GE81112 (▪), viomycin (□), edeine (▿), pactamycin (
), linezolid (⋄), sisomicin (○), and kasugamycin (▵). (C) Comparison of the effect of GE81112 and of other antibiotics on 027mRNA-directed S. cerevisiae translational systems in vitro: GE81112 (▪), viomycin (□), edeine (▿), pactamycin ( ), linezolid (⋄), sisomicin (○), and kasugamycin (▵). (D) Effect of GE81112 on in vitro bacterial (E. coli) (open symbols) and archaeal (S. sulfataricus) (closed symbols) translation of leadered 027mRNA (▴ and □) and leaderless Sui1mRNA (▴ and ▵). Further experimental details are given in Materials and Methods.
), linezolid (⋄), sisomicin (○), and kasugamycin (▵). (D) Effect of GE81112 on in vitro bacterial (E. coli) (open symbols) and archaeal (S. sulfataricus) (closed symbols) translation of leadered 027mRNA (▴ and □) and leaderless Sui1mRNA (▴ and ▵). Further experimental details are given in Materials and Methods.
Thus, the efficiency and selectivity of purified GE81112 in inhibiting mRNA translation in both prokaryotic (Fig. 2B) and S. cerevisiae (Fig. 2C) cell-free systems were compared to that of other antibiotics regarded or suspected to be translation initiation inhibitors (see Discussion). The results obtained demonstrate that, among all antibiotics tested, viomycin (IC50 = 0.6 μM) and GE81112 (IC50 = 0.9 μM) are the most effective inhibitors of the E. coli translational system (Fig. 2B), being more effective than pactamycin, edeine, sisomicin, kasugamycin, and linezolid. Furthermore, whereas most of the antibiotics tested, including GE81112, were ineffective on the eukaryotic translation system, both edeine and pactamycin were found to inhibit both prokaryotic and eukaryotic systems with comparable efficiencies. The IC50 of GE81112 in yeast (Fig. 2C) and HeLa cells (not shown) was found to be approximately two orders of magnitude higher than in the E. coli translational system. In addition to inhibiting protein synthesis in E. coli in vivo (Fig. 2A) and in vitro (Fig. 2B), GE81112 was found to be effective in vitro also toward mRNA translation in cell-free extracts derived from two clinical isolates of Pseudomonas aeruginosa having several antibiotic resistances (e.g., gentamycin and kanamycin) and from the Gram-positive Bacillus stearothermophilus. Indeed, the IC50 of GE81112 in the cell-free systems derived from these bacteria was found to be almost identical to that found in E. coli (data not shown). Furthermore, in contrast to what happens in cell-free translational systems derived from both lower and higher eukaryotic systems, the system derived from the archaeon Sulfolobus sulfataricus is inhibited by GE81112 with an IC50 similar (i.e., only ≈4 times higher) to that seen in the E. coli system. In addition, translation of a leaderless mRNA is inhibited in both systems with the level of inhibition somewhat higher in Archaea and somewhat lower in E. coli than that of a leadered mRNA (Fig. 2D). Taken together, these results indicate that GE81112 is a very potent, broad-spectrum, inhibitor of prokaryotic translation.
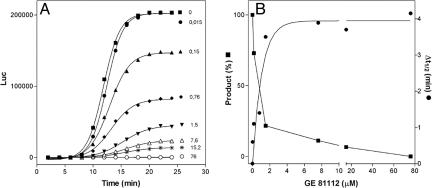
In the above experiments, translational inhibition by GE81112 was assessed by measuring the amount of protein synthesized after 30 min at 37°C. However, if enough time is allowed, even a translational system functioning at a reduced rate in the presence of an inhibitor could eventually yield an amount of product similar to that obtained in the noninhibited controls. Thus, the effect of the antibiotic was also tested on the kinetics of Renilla luciferase synthesis.
As seen from Fig. 3A, in the control samples the luciferase activity develops as a sigmoidal function within a rather narrow time interval after a lag period of ≈8 min (at 20°C), which is attributed to the time required for cotranslational folding of a polypeptide long enough to be endowed with enzymatic activity (9, 10). In the presence of increasing concentrations of GE81112, not only does the level of luciferase become lower (Fig. 3B) but also the lag becomes increasingly longer. In fact, the time at which 50% of the total luciferase activity is expressed increases from 11.84 min (in the absence of inhibitor) to 15.99 min in the presence of 76 μM GE81112, with a net Δt1/2 increase of ≥4 min (Fig. 3B). This lengthening of the lag is compatible with an interference of GE81112 with the early steps of translation.
Fig. 3.
Effect of GE81112 on in vitro Renilla luciferase synthesis. (A) Time course of in vitro synthesis of Renilla luciferase in the presence of the indicated concentrations (μM) of GE81112. (B) Level (▪) and Δt1/2, namely the time required for the synthesis of one-half the maximum level (•) of luciferase synthesized in the presence of the GE81112 concentrations indicated in the abscissa. The Δt1/2 was calculated by fitting the experimental points with the Boltzmann sigmoidal equation. The IC50 determined from this experiment was 0.8 μM. Additional experimental details are given in Materials and Methods.
To ascertain that inhibition by GE81112 is indeed due to inhibition of translation initiation, the effect of this drug was tested on various partial reactions of the initiation pathway. One of the earliest events occurring during translation initiation is the formation of a 30S initiation complex in which fMet-tRNA is bound to the P-site of the small (30S) ribosomal subunit in response to an initiation codon present in the translation initiation region of the mRNA (11, 12). As seen in Fig. 4A, increasing concentrations of GE81112 produce a strong inhibition of the formation of this complex; furthermore, the GE81112 dose–response curves obtained for mRNA translation and 30S initiation complex formation are virtually superimposable (Fig. 4A), suggesting that the antibiotic targets this step of translation and that inhibition of this function can account for all of the inhibitory activity displayed by GE81112 on mRNA translation. In light of these results, the efficiency of the GE81112 inhibition of fMet-tRNA binding to the 30S subunit was compared with that produced by other antibiotics reported to be “P-site inhibitors” such as edeine, pactamycin, kasugamycin (1–4, 13–15), and viomycin, which was tested because an old report indicated that, in addition to being a translocation inhibitor, viomycin is also able to inhibit fMet-tRNA binding to phage MS2 RNA-programmed 30S ribosomal subunits (16). The results demonstrate that GE81112, up to 1 μg/ml (i.e., ≈1.5 μM), is more active in inhibiting formation of 30S initiation complex than comparable concentrations of either pactamycin or edeine, whereas kasugamycin and viomycin, as expected, inhibit this ribosomal function little or not at all (Fig. 4B).
Fig. 4.
Effect of GE81112 on 30S initiation complex formation. (A and B) Comparison of the inhibition caused by the indicated concentrations of GE81112 on translational activity (▪) and on 30S initiation complex formation (▴) (A) and by the indicated concentrations of the following known or presumed P-site inhibitors (B): GE81112 (▪), viomycin (□), edeine (▿), kasugamycin (▵), and pactamycin ( ). (C) Effect of the order of addition of GE81112 with respect to the translation components. The indicated concentrations of GE81112 were preincubated (5 min at 37°C) with 30S ribosomal subunits before the addition of all other components of the translational machinery (▾) or added after 70S initiation complex formation and before the ingredients required for translation elongation (•). Additional experimental details are given in Materials and Methods.
). (C) Effect of the order of addition of GE81112 with respect to the translation components. The indicated concentrations of GE81112 were preincubated (5 min at 37°C) with 30S ribosomal subunits before the addition of all other components of the translational machinery (▾) or added after 70S initiation complex formation and before the ingredients required for translation elongation (•). Additional experimental details are given in Materials and Methods.
The data of Fig. 4A suggest that GE81112 inhibition occurs only once (i.e., at the initiation step) and is not reiterated during elongation. Consistent with this premise is the finding that the inhibitory activity of GE81112 is strongly reduced as the translational machinery proceeds from the early to the late stages of initiation. In fact, as seen in Fig. 4C, the IC50 of GE81112 is ≈15-fold lower (≈0.6 vs. 9 μM) if this antibiotic is preincubated with the 30S ribosomal subunit before the addition of all other components of the translational machinery or is given after the addition of the components required for 70S initiation complex formation but before addition of the components required for elongation.
As mentioned above, preliminary experiments indicated that GE81112 binds to the 30S ribosomal subunit with much higher affinity than to the 50S ribosomal subunit. This finding suggests that this antibiotic prevents P-site binding of fMet-tRNA by targeting the small ribosomal subunit, thereby hindering its function in various possible ways. However, other mechanisms could be responsible for the inhibition. In fact, GE81112 could inhibit 30S initiation complex formation by targeting either IF2 or the initiator fMet-tRNA and thereby interfering with the formation of the fMet-tRNA–IF2 binary complex, which is considered a precursor of the 30S initiation complex (11, 12, 17). Thus, experiments were carried out to clarify these points. Tests of IF2 protection of fMet-tRNA from alkaline hydrolysis (18) demonstrated that GE81112 does not affect fMet-tRNA–IF2 interaction (data not shown). Other experiments showed that also poly(U)-dependent binding of Ac-Phe-tRNA to 30S ribosomal subunit is inhibited by GE81112 (data not shown), indicating that this antibiotic does not recognize any particular structural facet of either the tRNAfMet molecule (17) or mRNA. On the contrary, addition of increasing amounts of 30S, but not 50S, ribosomal subunits was found to progressively relieve the translational inhibition caused by GE81112 (data not shown) and allowed us to establish that the 30S subunit is indeed the target of GE81112 inhibition, a conclusion fully supported by the topographical localization of its binding site (see below).
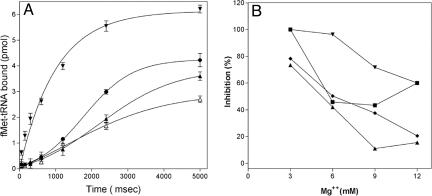
To gain additional insight into the actual mechanism by which GE81112 inhibits 30S initiation complex formation, its effect was studied under various experimental conditions. In the first experiment, the kinetics of fMet-tRNA binding to the 30S ribosomal subunits in the presence and absence of GE81112 was studied by using a rapid filtration device. As seen from Fig. 5A, the 30S initiation complex is formed rapidly in the absence of GE81112, reaching half saturation after ≈500 msec, and the experimental points can be fitted by a single-exponential equation. In contrast, the fMet-tRNA binding curves obtained in the presence of GE81112 are more complex, with the binding occurring after an initial lag period and yielding saturation levels that are lower with increasing antibiotic concentrations. Thus, GE81112 seems to determine an overall slower and less efficient ribosomal binding of fMet-tRNA, whereas the occurrence of the lag, qualitatively similar to that seen in the kinetics of luciferase synthesis (Fig. 3), suggests that initiator tRNA and GE81112 compete for binding to the 30S subunit. In the following experiment, fMet-tRNA binding to the 30S subunits was studied as a function of Mg2+ concentration in the presence and absence of different ribosomal ligands and of a fixed concentration (1.5 μM) of GE81112. Our findings demonstrate that the antibiotic inhibits fMet-tRNA binding under all conditions tested. However, the extent of inhibition strongly depends on conditions (i.e., Mg2+ concentration and presence or absence of initiation factors and mRNA) that influence the affinity of fMet-tRNA for the 30S subunit. Thus, GE81112 inhibition of 30S initiation complex formation is very strong (≈80%) at 3 mM Mg2+ but drops to <40% and <20% when the Mg2+ is increased to 9 and 12 mM, respectively. In the absence of mRNA or initiation factors, inhibition of fMet-tRNA binding is complete (≈100%) at 3 mM Mg2+ and, although somewhat relieved by Mg2+, remains quite high (≈45% and 75%) at ≥9 mM Mg2+. The only condition under which the substantial (≈80%) GE81112 inhibition is almost fully relieved by increasing the Mg2+ concentration is that in which IF3 was omitted (Fig. 5B). Taken together, these data indicate that GE81112 inhibits both coded and noncoded fMet-tRNA binding to the 30S subunit and regardless of whether the coded binding is assisted by the three initiation factors (Fig. 5B). Thus, it can be concluded that the main target of GE81112 inhibition is not the P-site decoding nor the functional interaction of the initiation factors with the ribosomal subunit and/or with its ligands but instead is the basal tRNA–30S subunit interaction. This conclusion is consistent with the observed effect of changes of Mg2+ concentration and other variables on the extent of GE81112 inhibition. It has been observed, in fact, that all conditions that determine an increased affinity of the ribosomal subunit for fMet-tRNA cause a reduction of the inhibitory power of the antibiotic, whereas the presence of IF3, whose capacity to increase the rates of formation and dissociation of the 30S initiation complex is well known (11, 12), seems to enhance the inhibition by GE81112.
Fig. 5.
Inhibition of 30S initiation complex formation by GE81112. (A) Kinetics of fMet-tRNA binding to 30S ribosomal subunits programmed with 022mRNA in the absence (▾) and in the presence of 0.25 μg/ml (0.37 μM) (•), 0.5 μg/ml (0.75 μM) (▴), and 1 μg/ml (1.5 μM) (▵) GE81112. (B) Codon dependence and initiation factor dependence of GE81112 inhibition of fMet-tRNA binding to 30S ribosomal subunits as a function of Mg2+ concentration. The experiment was performed in the presence of the indicated concentration of MgCl2 as described in Materials and Methods in a complete system (♦) or in systems in which IF1, IF2, and IF3 (▾), mRNA (▪), and IF3 (▴) were omitted.
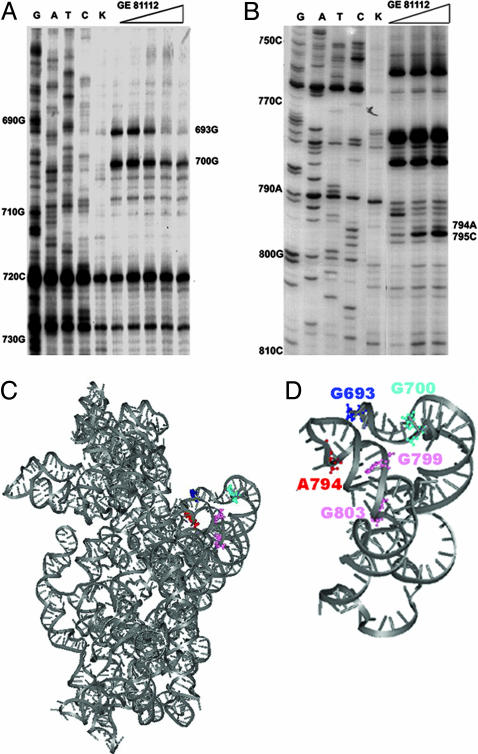
In light of the premise that the 30S ribosomal subunit is the target of GE81112 inhibition, chemical probing experiments of the 16S rRNA were performed in its presence to identify its ribosomal binding site. As seen from the primer extension analysis of the modified rRNA, GE81112 provides a substantial protection of G693 from kethoxal modification (Fig. 6A), exposes C795 to dimethyl sulfate (Fig. 6B), and causes smaller protections of G700 (Fig. 6A), A792, and C796 and somewhat increased reactivity of A780, A794, G799, and G803 (Fig. 6B). Some of these bases are the same as those protected by P-site-bound tRNA (19) and by other inhibitors such as edeine, kasugamycin, and pactamycin, which are traditionally considered P-site inhibitors (14, 15, 20). For instance, G693 is protected also by pactamycin and edeine, whereas A794, which is exposed by GE81112, is protected by kasugamycin and edeine (14, 20), and its mutation confers resistance to kasugamycin (21); C795, on the other hand, is exposed also by kasugamycin but is protected by pactamycin and edeine (14, 20, 21). The location of the bases affected by GE81112 are highlighted within the 3D structure of 16S rRNA (Fig. 6 C and D). Overall, these bases define a region between the platform and the E-site of the 30S subunit. In turn, this finding suggests that inhibition by GE81112 does not involve its direct binding to the P-site but probably entails an indirect mechanism of action (see Discussion).
Fig. 6.
Identification of the GE81112 target on the 30S ribosomal subunit. The 16S rRNA bases whose reactivity with kethoxal (A) and dimethyl sulfate (B) is affected by GE81112 were identified by primer extension analysis (8). The localization of these bases within the 3D structures of the entire E. coli 16S rRNA (C), and of the subunit-interface side of the platform (D) are also indicated (ref. 37, Protein Data Bank ID 1PNX).
Discussion
Protein synthesis is the fundamental biological process inhibited by the majority of known antibiotics. However, whereas elongation activities such as aminoacyl-tRNA binding and decoding in the ribosomal A-site, transpeptidation, and translocation are frequently found to be affected by antibiotics, albeit with different mechanisms, other functions such as translation initiation, termination, and aminoacylation are rarely or never found to be inhibited (1–4). Thus, the relevance and uniqueness of the present finding lie in the identification and characterization of the functional properties of GE81112, a tetrapeptide, which proved to be the most selective and effective inhibitor of prokaryotic 30S initiation complex formation known so far.
The mechanism of translation initiation is unique among the various steps of translation insofar as it involves the direct binding of fMet-tRNA in the ribosomal P-site through a process mediated by initiation factor IF2 (11, 12). However, unlike the case of elongation factors, which are directly targeted by inhibitors such as fusidic acid (EF-G) (22), kirromycin, pulvomycin, and the thiazolyl peptide GE2270A (EF-Tu) (23, 24), no known antibiotic is capable of interfering efficiently and specifically with the IF2–fMet-tRNA interaction. Furthermore, although it is known (and fully confirmed by the present study) that none of the few antibiotics that are considered “P-site inhibitors” (e.g., edeine, pactamycin, and kasugamycin, as reported in refs. 1–4) is either selective for prokaryotes or specific for a single molecular target, GE81112 was found to target specifically the 30S ribosomal subunit and to interfere exclusively with fMet-tRNA binding to the P-site; in fact, the dose–response curves of the inhibition of P-site tRNA binding and of translation inhibition are almost superimposable.
Unlike what happens in “elongation,” aminoacyl-tRNA–EF-Tu complexes, which enter into the A-site of the 70S ribosomes from the “shoulder” and “L7/L12 stalk” side of the 30S and 50S, respectively, some experimental evidence (25) suggests that the entry of fMet-tRNA into the P-site of the 30S subunit might occur from the platform side, possibly via the E-site, which is “empty” during the initiation phase of translation. The fairly extended lags in the kinetics of 30S initiation complex formation and luciferase synthesis observed in the presence of increasing concentrations of GE81112, which suggest a competition between the drug and the tRNA, the topographical localization of GE81112 near the 30S E-site, and the finding that inhibition occurs also with aminoacyl-tRNAs other than fMet-tRNA, whether coded or not coded, in the presence or absence of initiation factors, suggest that this drug does not inhibit the P-site directly. Instead, on the basis of the present results, it is tempting to speculate that the mechanism by which GE81112 inhibits fMet-tRNA binding to the P-site is indirect and consists of blocking (or interfering with) the path of fMet-tRNA binding (entry) to the small ribosomal subunit, before P-site decoding. Consistent with this premise is the finding that addition of GE81112 to the 30S subunits after that of mRNA and fMet-tRNA results in a much lower level of inhibition so that 70S initiation complex formation is much less sensitive to inhibition than 30S initiation complex formation. The fact that the binding site of this antibiotic is progressively obstructed as the ribosome becomes progressively committed to the elongation phase so that translating ribosomes (whose E-site is occupied by tRNA) are completely insensitive to the inhibition is also consistent with our premise. If GE81112 indeed interferes with the ribosomal entry of fMet-tRNA through the E-site, also other antibiotics binding at or near the E-site (edeine, kasugamycin, and pactamycin) could interfere with initiation complex formation by a similar yet less selective mechanism.
In fact, both literature survey and direct comparisons have shown that the efficiency and/or specificity of GE81112 is superior to that of all translation initiation inhibitors described so far. In fact, three of the antibiotics tested, namely linezolid, sisomicin, and, in contrast to an old claim (16), viomycin, were found to be completely ineffective in inhibiting translation initiation, whereas the other three (kasugamycin, pactamycin, and edeine) were found to be less effective or not prokaryotic-specific or both.
Also, the synthetic oxazolidinone antibiotic linezolid and the aminoglycoside sisomicin were found to be completely ineffective in inhibiting formation of 30S initiation complex. This finding is not entirely unexpected in the case of linezolid in light of the fact that, even though some reports indicate that this antibiotic could inhibit translation initiation, the concentration required to obtain this inhibition is so high that inhibition of initiation probably does not represent the primary mode of action (26, 27). Sisomicin, on the other hand, was reported to be the most effective molecule (IC50 ≈ 0.7 nM) within a group of aminoglycosides found to inhibit the IF2–fMet-tRNA interaction (28). However, in our hands this antibiotic proved to be a rather poor inhibitor of bacterial translation (Fig. 2B) and to have no effect whatsoever on 30S initiation complex formation and on fMet-tRNA–IF2 interaction (data not shown).
Other activity comparisons were made among GE81112 and kasugamycin, pactamycin, and edeine. Kasugamycin binds to the 30S subunit (and 70S ribosomes) with Kd = 2 × 10–5 M and is reported to enhance accuracy of protein synthesis (29) and to inhibit the initiation phase of translation without affecting elongation (30, 31), but unlike GE81112 (Fig. 2D), kasugamycin does not inhibit leaderless mRNA translation (32, 33). Kasugamycin was reported to destabilize 30S (but not 70S) initiation complexes, from which it removes prebound fMet-tRNA (34), but under our experimental conditions, kasugamycin inhibits prokaryotic protein synthesis with IC50 ≈ 30 μM, a value that is ≈40 times higher than that of GE81112 (Fig. 2B). Furthermore, kasugamycin was more than two orders of magnitude less effective than GE81112 in inhibiting 30S initiation complex formation (Fig. 4B). Thus, although some of the properties of kasugamycin are similar to those of GE81112, it is nevertheless a less efficient and, in light of its effect on eukaryotic phytopathogenic fungi (for a review see ref. 35), not prokaryotic-specific.
Finally, edeine and pactamycin, which are regarded as the most typical P-site inhibitors known so far, proved to be less effective translation initiation inhibitors than GE81112 and also not prokaryotic-specific. In fact, although edeine and pactamycin are nearly as effective as GE81112 in inhibiting bacterial translation in vitro (Fig. 2B), they also inhibit the eukaryotic system (Fig. 2C), in agreement with the report that they bind not only to prokaryotic 30S ribosomal subunits but also to eukaryotic 40S ribosomal subunits (1) and inhibit initiation complexes made with 40S subunits (13). Furthermore, in the direct comparison of their effect on 30S initiation complex formation (Fig. 4B), GE81112 proved to be more effective than both edeine and especially pactamycin, whose role as an initiation inhibitor has recently been questioned after a systematic analysis of its effects on each step of translation initiation and elongation (15). Finally, it should be mentioned that, unlike that of GE81112 (Fig. 2 A), the inhibitory activity of edeine is not selective for translation because this antibiotic clearly inhibits other cell functions (i.e., DNA synthesis) in addition to protein synthesis (36).
Acknowledgments
We are particularly grateful to Dr. Claudio Quarta for his support and encouragement and to our colleague, Prof. C. L. Pon, for her stimulating and valuable discussions and her generous help. This work was supported by European Commission Grant “Ribosome Inhibitors” (contract QLRT-2001-00892).
Author contributions: S.D. and C.O.G. designed research; L.B., A.F., A.L.T., and M.A. performed research; D.L. contributed new reagents/analytical tools; C.O.G. analyzed data; and A.F. and C.O.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: IF, initiation factor.
References
- 1.Gale, E. F., Cundliffe, E., Reynolds, P. E., Richmond, M. H. & Waring, M. J. (1981) in The Molecular Basis of Antibiotic Action (Wiley, London), pp. 278–379.
- 2.Walsh, C. (2003) Antibiotics: Actions, Origins, Resistance (Am. Soc. Microbiol., Washington, DC), pp. 50–69.
- 3.Cundliffe, E. (1990) in The Ribosome. Structure, Function & Evolution, eds. Hill, W. E. Dahlberg, A., Garrett, R. A., Moore, P. B., Schlessinger, D. & Warner, J. R. (Am. Soc. Microbiol., Washington, DC), pp. 479–490.
- 4.Wilson, D. N. (2004) in Protein Synthesis and Ribosome Structure, eds. Nierhaus, K. & Wilson, D. N. (Wiley-VSH, Weinheim, Germany), pp. 449–527.
- 5.La Teana, A., Pon, C. L. & Gualerzi, C. O. (1993) Proc. Natl. Acad. Sci. USA 90, 4161–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zubay, G. (1973) Annu. Rev. Genet. 7, 267–287. [DOI] [PubMed] [Google Scholar]
- 7.Brandi, A., Pietroni, P., Gualerzi, C. O. & Pon, C. L. (1996) Mol. Microbiol. 19, 231–240. [DOI] [PubMed] [Google Scholar]
- 8.Moazed, D. & Noller, H. F. (1986) Cell 47, 985–994. [DOI] [PubMed] [Google Scholar]
- 9.Kolb, V. A., Makeyev, E. V. & Spirin, A. S. (2000) J. Biol. Chem. 275, 16597–16601. [DOI] [PubMed] [Google Scholar]
- 10.Ferdorov, A. N. & Baldwin, T. O. (1995) Proc. Natl. Acad. Sci. USA 92, 1227–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gualerzi, C. O. & Pon, C. L. (1990) Biochemistry 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- 12.Gualerzi, C. O., Brandi, L., Caserta, E., Garofalo, C., Lammi, M., La Teana, A., Petrelli, D., Spurio, R., Tomsic, J. & Pon, C. L. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 363–376. [DOI] [PubMed] [Google Scholar]
- 13.Odom, O. W., Kramer, G., Henderson, A. B., Pinphanichakarn, P. & Hardesty, B. (1978) J. Biol. Chem. 253, 1807–1816. [PubMed] [Google Scholar]
- 14.Pioletti, M., Schlunzen, F., Harms, J., Zarivach, R., Gluhmann, M., Avila, H., Bashan, A., Bartels, H., Auerbach, T., Jacobi, C., et al. (2001) EMBO J. 20, 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinos, G., Wilson, D. N., Teraoka, Y., Szaflarski, W., Fucini, P., Kalpaxis, D. & Nierhaus, K. H. (2004) Mol. Cell 13, 113–124. [DOI] [PubMed] [Google Scholar]
- 16.Liou, Y. F. & Tanaka, N. (1976) Biochem. Biophys. Res. Commun. 71, 477–483. [DOI] [PubMed] [Google Scholar]
- 17.Rajbhandary, U. L. & Ming, C. (1995) in tRNA: Structure, Biosynthesis and Functions, eds. Söll, D. & Rajbhandary U. L. (Am. Soc. Microbiol. Press, Washington, DC), pp. 511–528.
- 18.Guenneugues, M., Caserta, E., Brandi, L., Spurio, R., Meunier, S., Pon, C. L., Boelens, R. & Gualerzi, C. O. (2000) EMBO J. 19, 5233–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noller, H. F., Hoang, L. & Fredrick, K. (2005) FEBS Lett. 579, 855–858. [DOI] [PubMed] [Google Scholar]
- 20.Woodcock, J., Moazed, D., Cannon, M., Davies J. & Noller, H. F. (1991) EMBO J. 10, 3099–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vila-Sanjurio, A., Squires, C. L. & Dahlberg, A. E. (1999) J. Mol. Biol. 293, 1–8. [DOI] [PubMed] [Google Scholar]
- 22.Bodley, J. W., Zieve, F. J. & Lin, L. (1970) J. Biol. Chem. 245, 5662–5667. [PubMed] [Google Scholar]
- 23.Parmeggiani, A. & Swart, G. W. (1985) Annu. Rev. Microbiol. 39, 557–577. [DOI] [PubMed] [Google Scholar]
- 24.Anborgh, P. H. & Parmeggiani, A. (1991) EMBO J. 10, 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirillov, S. V., Wower, J., Hixson, S. S. & Zimmermann, R. A. (2002) FEBS Lett. 514, 60–66. [DOI] [PubMed] [Google Scholar]
- 26.Burghardt, H., Schimz, K. L. & Mueller, M. (1998) FEBS Lett. 425, 40–44. [DOI] [PubMed] [Google Scholar]
- 27.Swaney, S. M., Aoki, H. & Ganoza, M. C. (1998) Antimicrob. Agents Chemother. 42, 3251–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans, J. M., Turner, B. A., Bowen, S., Ho, A. M., Sarver, R. W., Benson, E. & Parker, C. N. (2003) Bioorg. Med. Chem. Lett. 13, 993–996. [DOI] [PubMed] [Google Scholar]
- 29.van Buul, C. P., Visser, W. & van Knippenberg, P. H. (1984) FEBS Lett. 177, 119–124. [DOI] [PubMed] [Google Scholar]
- 30.Okuyama, A., Machiyama, N., Kinoshita, T. & Tanaka, N. (1971) Biochem. Biophys. Res. Commun. 43, 196–199. [DOI] [PubMed] [Google Scholar]
- 31.Kozak, M. & Nathans, D. (1972) J. Mol. Biol. 70, 41–55. [DOI] [PubMed] [Google Scholar]
- 32.Chin K., Shean, C. S. & Gottesman, M. E. (1993) J. Bacteriol. 175, 7471–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moll, I. & Blasi, U. (2002) Biochem. Biophys. Res. Commun. 297, 1021–1026. [DOI] [PubMed] [Google Scholar]
- 34.Poldermans, B., Goosen, N. & Van Knippenberg, P. H. (1979) J. Biol. Chem. 254, 9085–9089. [PubMed] [Google Scholar]
- 35.Tamamura, T. & Sato, K. (1999) Jpn. J. Antibiot. 52, 57–67. [PubMed] [Google Scholar]
- 36.Kurylo-Borowska, Z. (1964) Biochim. Biophys. Acta 87, 305–313. [DOI] [PubMed] [Google Scholar]
- 37.Vila-Sanjurjo, A., Ridgeway, W. K., Seymaner, V., Zhang, W., Santoso, S., Yu, K. & Cate, J. H. D. (2003) Proc. Natl. Acad. Sci. USA 100, 8682–8687. [DOI] [PMC free article] [PubMed] [Google Scholar]