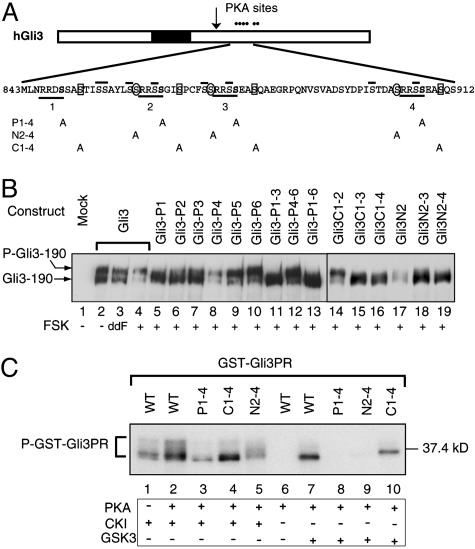
Fig. 2.
PKA-primed phosphorylation of Gli3 protein by CK1 and GSK3. (A)A schematic drawing of hGli3 protein. Six dots stand for PKA sites, and a filled box represents the zinc finger DNA-binding domain. An arrow indicates the approximate cleavage site. Underlined is the consensus sequence for PKA phosphorylation with S residues shown in bold. Numbers below the lines correspond to the designated names of the Gli3 mutant constructs. PKA-primed phosphorylation sites for GSK3 and CK1 are circled and boxed, respectively. Overlined are the secondary S residues that may potentially become GSK3 and CK1 phosphorylation sites. (B) Immunoblot analysis revealed phosphorylation of Gli3 and its mutants as measured by mobility shift of the proteins in transfected chicken limb bud cells after treatment with dideoxy FSK (ddFSK) and FSK. Proteins were separated by 5% SDS/PAGE. (C) PKA-primed phosphorylation of GST-Gli3PR (839–920 aa) and its mutants by CK1 and GSK3 in vitro. Affinity-purified GST-Gli3PR and its mutant proteins were first incubated with PKA catalytic subunit in the presence of nonradioactive ATP and subsequently with CK1 or GSK3 in the presence of [γ-32P]ATP. The phosphorylated proteins were resolved by SDS/PAGE and detected by autoradiography.