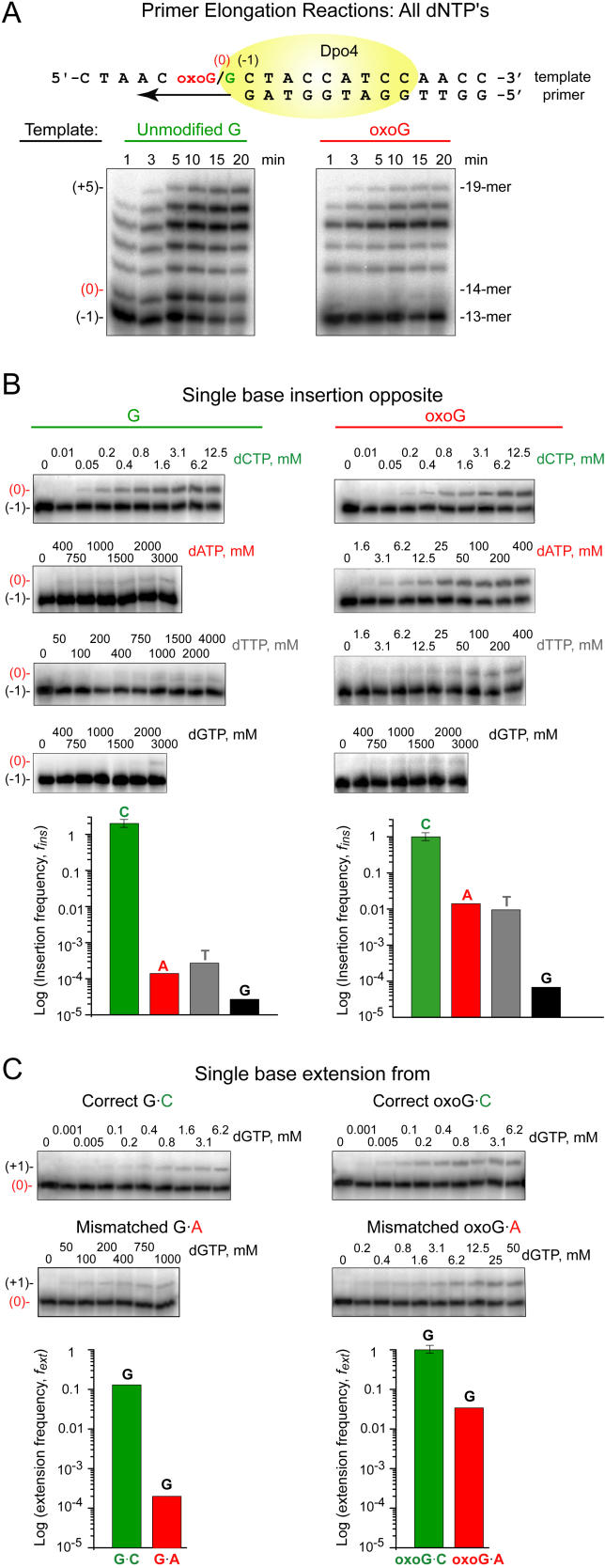
Figure 1. Fidelity and Efficiency of Base Incorporation on Unmodified G and oxoG-Modified Templates by Dpo4.
(A) Time course of 32P 5′-end-labeled 13-mer primer extension on 19-mer G (left panel) or oxoG (right panel) templates in the presence of all four dNTPs by Dpo4. The labeled (−1) position or 13-mer represents the 13-mer primer with the 3′ end positioned one base before the G or oxoG; the 14-mer extended to position (0) opposite G or oxoG; the 19-mer extended to position (+5) corresponds to full extension.
(B) Kinetics of single nucleotide insertion on G (left panel) and oxoG (right panel) templates by dCTP, dATP, dTTP and dGTP. The gels (top panels) show data as a function of dNTP concentration, while the plots (bottom panels) measure the log of the frequency of nucleotide insertion for each dNTP.
(C) Kinetics of single nucleotide extension from C and A opposite G (left panel) and oxoG (right panel) templates by the next correct nucleotide (dGTP). The gels (top panels) show data as a function of dGTP concentration, while the plots (bottom panels) measure the log of the frequency of nucleotide extension from G/oxoG•C and G/oxoG•A pairs. The primer extension data recorded under single-hit polymerization conditions (less than 20% of primer extended) were used for estimation of Michaelis-Menten parameters listed in Table 1.