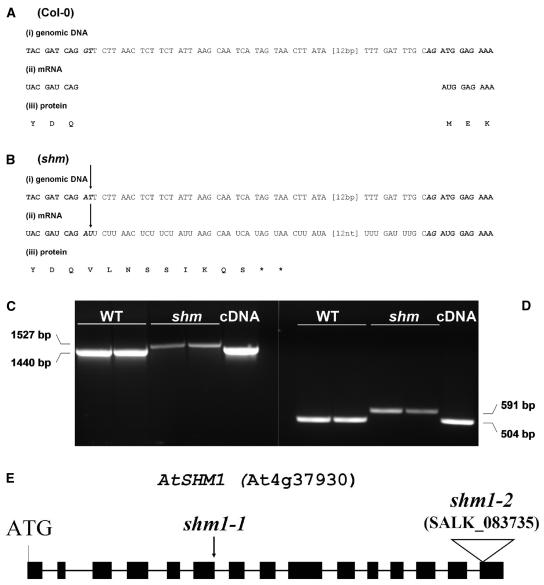
Figure 2.
Comparison between the genomic DNA (i), the mRNA (ii), and the derived protein sequence (iii) of wild-type (A) and shm1-1 mutants (B). Only the sequence between the end of exon 6 (start-ATG + 1,272 bp) and the beginning of exon 7 (start-ATG + 1,376 bp) is shown. Bold, Exon; italics, intron; bold italics, conserved splice motif. The arrows indicate the point mutation in shm1-1. Asterisk (*), Translational stop. C, RT-PCR products from wild-type and shm1-1 mRNA obtained with primers prSHM4 and prSHM9 (see Table I). D, Primers prSHM5 and prSHM6 (see Table I). Two independent RNA preparations of each line were taken for the RT-PCR reactions. From left to right, Two lanes wild-type (Col-0); two lanes shm1-1 mutant; one lane cDNA. The RT-PCR fragment sizes are indicated beside the image. Plasmid containing the SHM1 expressed sequence tag clone 148C5T7 was used as a positive control in the PCR reaction. E, Cartoon showing AtSHM1 gene structure and the shm1-2 T-DNA insertion site in the last of the 15 SHM1 exons. As a reference, the point mutation in shm1-1 is indicated by an arrowhead.