Abstract
As part of the embryo maturation process, orthodox seeds undergo a developmentally regulated dehydration period. The LATE EMBRYOGENESIS ABUNDANT (LEA) genes encode a large and diverse family of proteins expressed during this time. Many hypothesize that LEA proteins act by mitigating water loss and maintaining cellular stability within the desiccated seed, although the mechanisms of their actions remain largely unknown. The model plant Arabidopsis (Arabidopsis thaliana) contains two genes belonging to the group 1 LEA family, ATEM1 and ATEM6, and knockout mutations in these genes are being sought as a means to better understand group 1 LEA protein function during embryo maturation. We have identified a T-DNA insertion allele of the ATEM6 gene in which the T-DNA is present just downstream of the protein coding region. While this gene is transcriptionally active and encodes a wild-type protein, there is no detectable ATEM6 protein in mature seeds. Mutant seeds display premature seed dehydration and maturation at the distal end of siliques, demonstrating that this protein is required for normal seed development. We propose that one function for group 1 LEA proteins in seed development is to buffer the water loss that occurs during embryo maturation and that loss of ATEM6 expression results in the mutant phenotype.
Seeds are remarkable structures that serve as the connection between successive generations of plants. As part of the normal developmental program of orthodox seeds, most cellular water is lost during the latter stages of embryo maturation and many seeds dehydrate to as low as 5% of their original water content. Despite the low level of retained water, seed embryos remain viable and mature, and dry seeds readily germinate under appropriate environmental conditions. Therefore, plants must have evolved a mechanism that allows cells to tolerate the low water potential found in dry seeds.
The LATE EMBRYOGENESIS ABUNDANT (LEA) genes are expressed, as the name implies, late in embryo maturation and during the developmentally regulated period of dehydration at the end of seed development. LEA proteins are thought to play an important role in the maturation process, and studies in barley (Hordeum vulgare) have demonstrated that accumulation of LEA proteins in embryos is correlated with the acquisition of desiccation tolerance (Bartels et al., 1988). This correlation suggests that these proteins are an integral part of one or more tolerance mechanisms. Many LEA genes can also be induced by the application of abscisic acid and by various kinds of water stress in both reproductive and vegetative tissues (Berge et al., 1989; Bostock and Quatrano, 1992; Wilhelm and Thomashow, 1993; Bies et al., 1998). In addition, there is at least one example of a constitutively expressed LEA gene (Welin et al., 1994).
LEA genes are found throughout higher plants, including monocots, dicots, and gymnosperms, and differences in temporal expression pattern and sequence similarity among the encoded proteins have allowed them to be subdivided into groups (Dure et al., 1989; Cuming, 1999; Wise, 2003). Based largely on predictions of regions of secondary structure, the different groups have been proposed to contribute in one way or another to the maintenance of viability. For example, the group 1 and group 4 LEA proteins are predicted to exist largely as random coils (McCubbin et al., 1985; Soulages et al., 2002). With a high degree of flexibility, these proteins could interact with a variety of cellular components, including other proteins, and contribute to stability either by sharing their hydration shell of water or by using their own hydroxylated amino acids to serve as a replacement for water as it is lost from the cell (water replacement hypothesis). Group 2 and group 3 LEA proteins have also been hypothesized to contribute to cellular stability through the sequestration of ions and membrane stabilization, respectively (Dure, 1993).
The group 1 LEA genes, also called the Em (Thompson and Lane, 1980) or D-19 group (Baker et al., 1988) genes, have been identified in a wide range of plants and have the highest degree of sequence homology at both the nucleotide and amino acid sequence levels (Dure et al., 1989). Induction of these genes appears to coincide with an elevation in embryonic abscisic acid levels and expression continues late into development. These genes are also expressed early during the germination process from stored mRNA (Williamson and Quatrano, 1988; Morris et al., 1990). Group 1 LEA protein primary structure is typified by the presence of nearly 20 mol% Gly residues and a preponderance of charged and hydroxylated amino acids. All group 1 LEA proteins also contain a highly conserved stretch of 20 amino acids that may be tandemly repeated up to four times. This tandem arrangement of the repeat has been suggested to be the result of gene duplication followed by recombination or deletion (Stacy et al., 1995). Group 1 LEA proteins have been isolated from several plant species and characterized biochemically and structurally (McCubbin et al., 1985; Russouw et al., 1995, 1997). These studies have revealed that the group 1 LEA proteins exist predominantly as random coils in solution and that they have an unusually large sphere of hydration. The random coil configuration appears to be unchanged after heating to 80°C for 10 min (Russouw et al., 1997).
There are two group 1 LEA genes, ATEM1 and ATEM6, in the genome of the model plant Arabidopsis (Arabidopsis thaliana), and these genes are differentially expressed both temporally and spatially during embryo maturation. ATEM1 mRNA is expressed predominantly in provascular tissues with the strongest expression in the root tip (Vicient et al., 2000) and ATEM1 expression initiates approximately 2 d prior to ATEM6 (Bies et al., 1998). ATEM6 mRNA is expressed in essentially all regions of the embryo with the strongest expression in the shoot apical meristem and provascular tissue (Vicient et al., 2000). These two genes encode similar proteins that differ mainly by the number of repeats in the conserved 20-amino acid motif (four copies in ATEM1 and one copy in ATEM6).
To study the in planta function of the ATEM proteins during embryo maturation, a population of more than 100,000 T-DNA-mutagenized Arabidopsis lines maintained by Syngenta (Sessions et al., 2002) was screened for potential ATEM1 and ATEM6 gene knockout lines. Here we report the molecular and phenotypic characterization of one insertional knockout mutation in the ATEM6 gene and the location of a number of non-LEA insertions identified during the course of this study.
RESULTS
Approximately 3 kb of Arabidopsis genomic DNA sequence containing the ATEM6 locus (At2g40170) was provided to the Torrey Mesa Research Institute for a search of the Syngenta T-DNA insertional mutation database (Sessions et al., 2002). The results of the BLAST search were analyzed and the sites of two potential insertions within this gene were identified (Table I). One plant line, Garlic_413_E08, contains T-DNA from pCSA110 (McElver et al., 2001) and was predicted to contain an insertion at position 16,786,882 on chromosome 2. This location is within exon 2 of the ATEM6 coding region and 10 bp 5′ of the TAA translational stop codon. The insertion in the other line, Garlic_587_G09, contains T-DNA from pCSA104 (McElver et al., 2001) and was predicted to be located at position 16,787,092 on chromosome 2. This location is within the single intron of ATEM6.
Table I.
Plant lines
| Garlic Line No. | Abbreviated No. | Ti Plasmid | Insertion Site in ATEM6 Putative/Actual |
|---|---|---|---|
| Garlic_413_E08 | 6.413 | pCSA110 | 16,786,882/6,786,867 |
| Garlic_587_G09 | 6.587 | pCSA104 | 16,787,092/No insert |
Seeds for the two plant lines were obtained, grown in soil, and both seed and vegetative tissue were harvested from the initial planting. Genomic DNA was extracted from the bulked plant tissue of each line and subjected to Southern-blot analysis using T-DNA-specific (pBluescript [pBS]) and ATEM6 gene-specific (3′-untranslated region [UTR]) probes after digestion with an enzyme that cleaves once near the center of the T-DNA. While both lines displayed multiple bands when hybridized with the T-DNA probe (pBS), indicating the presence of multiple insertion sites, only the Garlic_413_E08 line displayed a band other than that found in wild-type Columbia genomic DNA when hybridized with the ATEM6 gene-specific 3′-UTR probe (data not shown). This demonstrates that the Garlic_413_E08 line contains a T-DNA in or near ATEM6 and that the Garlic_587_G09 line does not. This Garlic_413_E08 plant line will hereafter be referred to as 6.413, as a reference to the ATEM6 gene and the Garlic plant line number supplied by Syngenta.
Southern-Blot Analysis
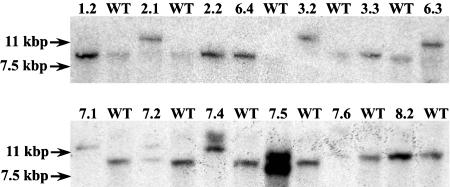
Southern hybridization assays using an ATEM6 3′-UTR gene-specific probe were performed to identify individual plants containing the T-DNA insertion identified above. Digestion of genomic DNA with BamHI should produce an approximately 11.4-kb DNA fragment containing both the pBS portion of the T-DNA and the ATEM6 3′-UTR region for the T-DNA insertion allele (Fig. 1A), whereas the wild-type ATEM6 allele should produce an 8,470-bp fragment, a difference of approximately 3.0 kb. Figure 2 shows that plants grown from the original bulked seed are segregating for the insertion and that individual plants homozygous for the wild-type ATEM6 allele (e.g. lines 1.2, 2.2, and 6.4), homozygous for the T-DNA insertion allele (e.g. lines 2.1, 3.2, and 8.2), and heterozygotes (e.g. line 7.2) are clearly distinguishable. Hybridization of the same membranes with a pBS probe demonstrated that all individuals contained multiple T-DNA inserts (data not shown). Plants homozygous for the ATEM6 insertion allele were backcrossed into wild-type Columbia and the resulting seedlings screened using the Southern-blot analysis described above to identify plants with only the ATEM6-associated T-DNA. These heterozygous plant lines were allowed to self and plant lines homozygous for the insertion allele were subsequently identified.
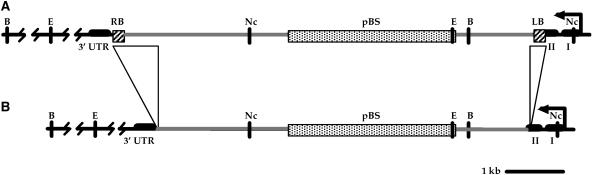
Figure 1.
Schematic of pCSA110 T-DNA in ATEM6. The T-DNA region is shown as a simple, single insertion in ATEM6 genomic DNA (A) and the actual structure as determined by plasmid rescue and sequence analysis of the RB/ATEM6 and LB/ATEM6 junctions (B). Genomic DNA is black. The T-DNA, bounded by the RB and LB sequences (hatched), is gray, and the pBS portion of the T-DNA is stippled. Exons of the ATEM6 gene are shown as thick bars (I, II, and 3′-UTR). Note that the 3′-UTR region of exon II is physically separated from the remainder of the gene by the T-DNA insertion. Deletions of T-DNA identified by sequence analysis are underlined in A and absent in B. Restriction enzyme sites for EcoRI (E), NcoI (Nc), and BamHI (B) are indicated.
Figure 2.
Southern-blot analysis of individual 6.413 mutant plants. A 10-μg sample of genomic DNA was digested with BamHI, fragmented on a 0.7% agarose gel, transferred to nylon membrane, and hybridized with an ATEM6 gene-specific 3′-UTR probe. WT, Wild type; numbers indicate laboratory plant line numbers. Molecular marker sizes are indicated to the left.
Plasmid Rescue
If the T-DNA present in the 6.413 plant line represents a simple, single insertion, as diagrammed in Figure 1A, the insert should be bounded by the left border (LB) and right border (RB) regions and it should be possible to PCR amplify boundary fragments using LB or RB and ATEM6 gene-specific primers. However, initial attempts at PCR failed to produce any products, suggesting that the DNA at the site of the insertions may have undergone some rearrangement, a relatively frequent observation for T-DNA insertions (Castle et al., 1993; Tax and Vernon, 2001). From the sequences for the plasmid pCSA110 provided by Syngenta, there is an EcoRI site very near one end of the pBS sequence and an NcoI site in the T-DNA adjacent to the pBS sequences at the end opposite from the EcoRI site (Fig. 1A). Therefore, a plasmid rescue strategy was used to recover border sequences flanking the T-DNA insert (Feldmann, 1992).
The EcoRI experiment to define RB junctions with genomic DNA rescued two plasmids (6.413-1E and 6.413-2E). When plasmid DNA from the two rescued colonies was extracted and run on an agarose gel for size determination, the two linearized plasmids appeared to be approximately the same size (approximately 7,500 bp). However, digestion with NcoI resulted in different banding patterns, demonstrating that these two plasmids are unique (data not shown). Sequence analysis of plasmid 6.413-1E indicated that this T-DNA insert was not within the ATEM6 gene locus, but rather at position 16,474,005 on chromosome 1 in the 5′-LB to RB-3′ orientation (with respect to the 5′ to 3′ orientation of the chromosome sequence; Table II). This insertion is most closely linked to locus At3g11860 that encodes a hypothetical protein. In addition, our data show that 189 bp of the RB sequence have been deleted. Sequence analysis of plasmid 6.413-2E confirmed the insertion of a T-DNA in a 5′-RB to LB-3′ orientation (with respect to the 5′ to 3′ orientation of the chromosome sequence) with its RB/ATEM6 junction at position 16,786,867 on chromosome 2 (Fig. 1B; Table II). The sequence analysis further revealed a 767-bp deletion at the RB of the T-DNA immediately 5′ to the insert location.
Table II.
Chromosomal location of T-DNAs in putative ATEM6 insertion lines
| Garlic Line No. | Chromosome | Locationa | Nearest Gene (AGI No.) | Function |
|---|---|---|---|---|
| 6.413 | 1 | 16,474,005 | At1g43680 | Hypothetical protein (function unknown) |
| 2 | 16,786,867 | At2g40170 | Putative desiccation tolerance protein | |
| 3 | 19,134,716 | At3g51560 | Disease resistance protein | |
| 6.587 | 2 | 19,636,488 | At2g47970 | NPL4 family protein |
| 3 | 11,810,368 | At3g30180 | Putative cytochrome P450 |
Nucleotide coordinates are as determined by BLAST search at http://tigrblast.tigr.org/er-blast/index.cgi?project=ath1.
The NcoI rescue experiment yielded nine plasmids (6.413-1N to 6.413-9N). Sequence analysis of one of these plasmids, 6.413-2N, indicated that this T-DNA insert was not within the ATEM6 gene locus, but rather at position 19,134,716 on chromosome 3 in the 5′-LB to RB-3′ orientation (with respect to the 5′ to 3′ orientation of the chromosome sequence; Table II). This insertion is most closely linked to locus At3g51560 that encodes a disease resistance protein of the Toll interleukin receptor-nucleotide-binding site-Leu-rich repeat class. The remaining eight plasmids, 6.413-1N and 6.413-3N to 6.413-9N, defined the ATEM6/LB junction at position 16,786,868 on chromosome 2 (Fig. 1B; Table II). Sequence data further indicated a deletion of 267 bp in the T-DNA LB region directly adjacent to the insert location. Therefore, this T-DNA insertion is located 15 bp 3′ of the predicted insertion site as determined by the initial BLAST search or 2 bp downstream of the TAA stop codon in ATEM6 (i.e. outside the ATEM6 open reading frame). Whereas there is deletion of T-DNA sequences at both ends of the insert, there is no accompanying deletion of ATEM6 sequences. As such, the ATEM6 open reading frame in exon 2 remains intact and still encodes a normal protein. The deletions in both the T-DNA LB and RB regions explain our earlier inability to PCR amplify products from these locations using Syngenta primers in conjunction with ATEM6 gene-specific primers. Furthermore, this suggests that the deletions occurred after the sequence information identified by BLAST search was generated for the Syngenta database (Sessions et al., 2002). Hereafter, the plant lines containing the T-DNA insertion in the ATEM6 locus will be referred to as atem6-1. Two additional inserts identified in the Garlic_587_G09 line, including nearest gene and putative function, are also shown in Table II.
Chromosome 2 Mapping
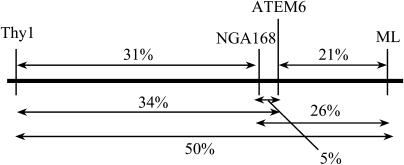
The plasmid rescue results described above indicated that there were deletions in both borders of the atem6-1 T-DNA. Although sequence analysis of the regions directly adjacent to the insertion did not indicate any chromosomal alterations, it remained possible that other aberrations on chromosome 2 had occurred. Therefore, one simple sequence-length polymorphism (NGA168) and two cleaved-amplified polymorphic sequence (Thy1 and ML) markers were chosen to map the large arm of chromosome 2. Thy1 is located close to the centromere, NGA168 is tightly linked to ATEM6, and ML is located close to the telomere. All markers mapped to chromosome 2 and in the correct order as compared to ATEM6 and to each other, indicating that there were no gross chromosomal abnormalities associated with the atem6-1 insertion (Fig. 3).
Figure 3.
Mapping of chromosome 2. Schematic of map positions of the Thy1, NGA168, and ML markers and the ATEM6 locus. The genetic distance was derived from recombination analysis of 100 individual F2 plants. Distances are given as percentage recombination.
Reverse Transcription-PCR and RNA Gel-Blot Analysis of Seeds
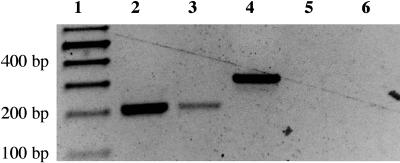
Sequence analysis from the plasmid rescue experiments demonstrated that the atem6-1 T-DNA insertion occurred 2 bp outside of the ATEM6 open reading frame. Since both the promoter and coding regions of the mutant allele remained intact, it could be possible for this gene to express an mRNA that encodes a wild-type protein. It was of interest, therefore, to determine whether this mutant allele expresses an mRNA. Total RNA extracted from mature wild-type seeds and from mature seeds homozygous for the atem6-1 allele was subjected to reverse transcription (RT)-PCR. The RT reaction was performed using an oligo(dT)17 primer containing a V clamp at the 3′ end and a 20-bp 5′ extension (Table III). Subsequent PCR was performed using primers designed to the ATEM6 first and second exons such that a 198-bp product should be amplified from ATEM6-derived mRNA for both the wild-type and atem6-1 mutant allele. Wild-type Columbia DNA was subjected to PCR in a separate reaction as a control for contaminating genomic DNA. Because the PCR primers flank the single intron in ATEM6, a product derived from a DNA template should be 303 bp. The generation of a 198-bp product from both wild-type atem6-1 mutant seeds demonstrates that an ATEM6-derived mRNA is actively transcribed in the mutant (Fig. 4).
Table III.
Sequencing primers
| Primer Name | Primer Sequence (5′→3′) |
|---|---|
| T7 promoter | GTAATACGACTCACTATAGGG |
| T3 promoter | AATTAACCCTCACTAAAGGG |
| QRB2 | GGGGTCATCTATGTTACTAGATCGGGAATTGA |
| QLB2 | GCTTCCTATTATATCTTCCCAAATTACCAATACA |
| AJM-LB1 | GGTGTTTTGGTTTTTTCTTGTGGCCGTC |
| ATEM6-F | CACAAATAACTTGCCTTCGTAAGAATCAC |
| ATEM6-R | CCGATCCAGTAATCATTCCTAATTCAATC |
| ATEM6 gene-specific 1 | CACACATCACGATTGATCCAACACTATGTTGAG |
| ATEM6 gene-specific 2 | CTAAAGAGCCATGGCGTCTCAACAAGAG |
| ATEM6 gene-specific 3 | CTTTGGAACCTTCCAAATTTCACAC |
| Tailed oligo(dT) | GCAAGCTTCGAGTGAATTCGT17V |
| ATEM6 RT-PCR forward | CATGGCGTCTCAACAAGAGA |
| ATEM6 RT-PCR forward | GTCTCCGGTGCTAAGACCAC |
| Tail alone | GCAAGCTTCGAGTGAATTCG |
| NptII forward | TCATTTCGAACCCCAGAGTC |
| NptII reverse | GCGTTCAAAAGTCGCCTAAAG |
| Thy1-forward | AACCGCCATTTTCATTTCTATC |
| Thy1-reverse | GGCGACCTTGGACCTGTATACG |
| NGA168-forward | GAGGACATGTATAGGAGCCTCG |
| NGA168-reverse | TCGTCTACTGCACTGCCG |
| ML-forward | CGGAAACACGAAGCTGATGAGTTGGG |
| ML-reverse | CGAGAACAAAATGTGTACGGTGTG |
Figure 4.
RT-PCR of wild-type and atem6-1 RNA. Total RNA extracted from 10 seeds per sample was reverse transcribed using a tailed oligo(dT) primer and subsequently subjected to PCR using ATEM6 exon-specific primers. A portion of the RT-PCR reaction was run on a 1.2% agarose gel. Note this is not a quantitative measure of mRNA levels. Lane 1, RNA markers (sizes shown to the left); lane 2, wild type; lane 3, atem6-1 mutant; lane 4, ATEM6 genomic clone; and lanes 5 and 6, no RT controls for wild type and atem6-1 mutants, respectively.
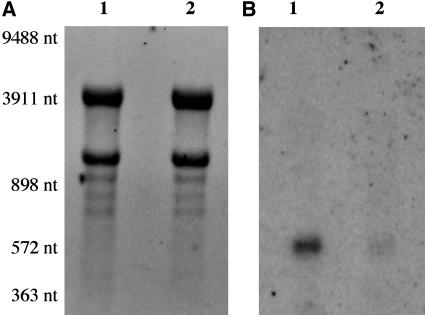
RNA gel-blot analysis of total RNA samples was then used to determine the level of expression and size of the wild-type and mutant transcripts. Previous cDNA characterization has shown that wild-type ATEM6 transcripts are approximately 540 and 640 nucleotides in length prior to polyadenylation (Gaubier et al., 1993). Because of the location of the T-DNA insertion in the mutant, the 3′-UTR region is physically separated from the remainder of the ATEM6 gene. Therefore, membranes were hybridized using an ATEM6 5′-UTR probe. As shown in Figure 5, the ATEM6 message is readily detected in wild-type mature seed RNA and is of a size consistent with the earlier study. Seeds homozygous for the mutant atem6-1 allele, however, contain significantly lower levels of a slightly smaller transcript, approximately 12% of that found in wild type.
Figure 5.
RNA gel-blot analysis of wild-type and atem6-1 RNA. Total RNA (10 μg) from mature seeds was separated on a 1.2% agarose formaldehyde gel and transferred to nylon membrane (A) and hybridized with an ATEM6 gene-specific 5′-UTR probe (B). Lane 1, atem6-1 mutant; lane 2, wild type. RNA Mr marker sizes are indicated to the left.
To determine the 3′ end of the mutant mRNA and to ensure that the seed from which the RNA was extracted was not contaminated with wild type, the RT reaction of atem6-1 mature seed RNA from above was used for 3′-RACE. The 5′ exon 1 RT-PCR primer was used in combination with a 3′ primer representing the 5′ extension on the RT primer (Table III). The products were cloned as blunt-ended fragments into pBS after treatment with Klenow fragment and T4 DNA kinase. Sequence analysis of eight clones identified two classes of transcripts from the atem6-1 mutant allele that are polyadenylated at 103 and 114 nucleotides after entering the T-DNA insert. None of the clones contained any wild-type ATEM6 3′-UTR sequences except the 2 bp that separate the T-DNA insertion from the open reading frame.
Immunoblot Analysis
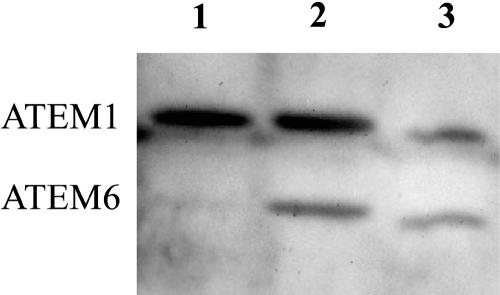
Since the mutant atem6-1 seeds express a mature ATEM6-derived mRNA, albeit at a significantly lower level than that found in wild type, it was of interest to determine whether these seeds also translate that mRNA into ATEM6 protein. Protein was isolated from mature seeds of a single silique from wild-type and homozygous atem6-1 plants as well as from a silique of a backcross of a homozygous atem6-1 mutant plant to wild type (i.e. heterozygous seed). The extracts were separated on a 15% Tris-Gly gel, transferred to a polyvinylidene difluoride (PVDF) membrane, and ATEM proteins were detected using a polyclonal rabbit anti-wheat Em protein antibody. Figure 6 shows that this antibody detects both ATEM6 and ATEM1 proteins in wild-type seed extracts. ATEM6 protein is also readily detected in the heterozygous atem6-1 seed extracts, but is essentially undetectable in homozygous atem6-1 mutant seed extracts. Therefore, although there is an actively transcribed message present in mature homozygous mutant atem6-1 seeds, there is no detectable ATEM6 protein.
Figure 6.
Immunoblot analysis of wild-type and atem6-1 protein. Total protein extracted from seeds from single siliques was separated on a 15% SDS-polyacrylamide gel and transferred to PVDF membrane. ATEM proteins were detected with an anti-wheat Em protein antibody. Note this is not a quantitative measure of protein levels. Lane 1, atem6-1 mutant; lane 2, ATEM6/atem6-1 heterozygote; lane 3, wild type. ATEM1 and ATEM6 proteins are indicated to the left.
Phenotypic Analysis of atem6-1 Mutant Plants
ATEM6 mRNA and ATEM6 protein are normally expressed only in embryonic tissues and only during the final stages of seed maturation (Gaubier et al., 1993; Bies et al., 1998; Vicient et al., 2000). As expected, the atem6-1 mutants display no abnormalities in vegetative growth and the transition from vegetative to reproductive development is in no way different from wild type. The molecular analyses above clearly show that the atem6-1 insertion line does express an mRNA for ATEM6, but there is little or no ATEM6 protein present in mature seeds. The ability to obtain viable homozygous atem6-1 mutant seeds that readily germinate, however, demonstrates that this mutation is not lethal and mature dry seeds from homozygous mutant plants are visibly indistinguishable from wild type.
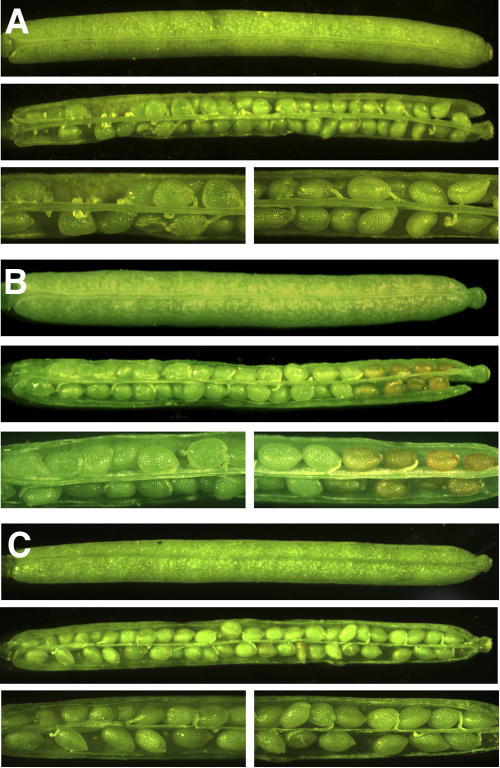
Under the growth conditions used in this study, silique and seed development was complete with mature dry seeds being present 16 to 17 d after flowering (daf). For this study, the phenotype of siliques ranging in age from 7 through 17 daf was examined. When homozygous atem6-1 mutant plants and wild-type Columbia plants were grown in parallel, it was observed that seed and silique development is also indistinguishable through about 12 daf. After that period, however, there is a visible difference between mutant and wild type and, at 13 and 14 daf, seeds at the distal end of siliques of mutant plants begin to dehydrate prematurely (Fig. 7, A and B). In addition, the distal ends of the mutant siliques also dehydrate early and, in the most severe cases, split open prior to any dehydration of seeds or siliques at the proximal ends.
Figure 7.
Phenotype of wild-type (A), atem6-1 mutant (B), and ATEM6-C (complementation; C) seeds and siliques. Siliques were removed from the plants at 13 daf and examined under a dissecting microscope. For each genotype, top image represents a whole silique; middle image is the same silique after opening; and bottom image is a closer view of the plant proximal (left) and plant distal (right) ends of the same silique.
To verify that earlier stages of seed development are not affected in this mutant, seeds were dissected from siliques at 8 to 15 daf, cleared using Hoyer's solution (Liu and Meinke, 1998), and visualized by Nomarski optics. There was no obvious difference between mutant and wild-type seed development through 12 daf (data not shown). When the phenotype was visible (13–14 daf), embryos at the base of the silique were visually as developed as those at the tip, the only difference being the premature dehydration of seeds at the distal end. Therefore, development of mutant embryos appears to be synchronous along the length of the silique, whereas the onset of dehydration is not. Consistent with expression only in embryonic tissues and only during the maturation phase of embryo development, growth at all other stages and in all other tissues appears indistinguishable from wild type (data not shown).
atem6-1 Complementation
The wild-type ATEM6 allele was reintroduced into homozygous atem6-1 mutant plants by direct T-DNA transformation using a 3,002-bp genomic clone. PCR was used to verify that the plants contained both the atem6-1 mutant allele and the ATEM6 complementation construct. Phenotypic characterization (as described above) indicated that the presence of the ATEM6 complementation construct is sufficient to rescue the phenotype of the atem6-1 mutant allele (Fig. 7C). Whereas mutant siliques exhibited premature desiccation of the seeds at the tip, wild-type and the atem6-1-complemented seeds desiccated synchronously along the length of the silique.
DISCUSSION
During normal development, the Arabidopsis genes ATEM1 and ATEM6 are expressed only in embryos. The temporal expression pattern of these two genes is slightly different, with ATEM1 induction preceding ATEM6 by about 2 d. Spatial expression patterns also differ with ATEM1 expressed predominantly in provascular tissue and the root meristem, whereas ATEM6 is expressed throughout the embryo with greater expression in provascular tissue and the shoot meristem (Vicient et al., 2000). Therefore, the proteins encoded by these genes are not functionally redundant in seed maturation.
By BLAST search of a T-DNA insertional mutant population, we identified a putative mutation in the ATEM6 gene. Molecular characterization of the insertion revealed that the T-DNA had inserted just downstream of the TAA stop codon for the open reading frame. Insertion at this location results in retention of an intact 5′ flanking region and an unaltered protein coding capability. Barring the involvement of downstream regulatory elements, one would predict that this insertion allele should exhibit normal transcriptional activity. Indeed, our results from RT-PCR demonstrate that this gene is transcribed and spliced in maturing seeds and that the transcripts are polyadenylated, at least to some extent. As evidenced by RNA gel-blot analysis, however, the transcript levels appear significantly reduced in dry seed.
The lower level of mRNA expression in the mutant could be explained in several ways. First, the onset of expression could be delayed or the rate of transcription could be lower. Although we think this is unlikely, we are currently investigating temporal regulation of ATEM6 in the atem6-1 mutant to determine whether it is comparable to that of wild-type plants. An alternative explanation, and the one we favor, is that abnormal transcription termination and/or polyadenylation of the mRNA is responsible for the decrease in mature seed mRNA levels. Our 3′-RACE data show that transcripts from the atem6-1 insertion allele are polyadenylated, at least to some extent, and terminate at two different places in the T-DNA. This suggests the use of cryptic or fortuitous termination signals present in the T-DNA sequences resulting in the production of a chimeric RNA that may have an altered stability with respect to wild type. In any case, the normal ATEM6 termination and polyadenylation signals will not be present as the T-DNA insert physically separates the normal 3′ region for this gene from the remainder by over 7.5 kb. The likelihood that a conventional termination/polyadenylation process is functioning, particularly one associated with a gene that is normally expressed to high levels, is remote.
Our data further demonstrate that, although the T-DNA insert is located outside the ATEM6 coding region of the gene, there is little or no ATEM6 protein present in mature seeds. This is not due to an inability to detect ATEM proteins in extracts from cells that contain the insertion allele because both ATEM6 and ATEM1 proteins are seen in extracts from heterozygotes. While the antibody used is polyclonal, it is likely that similar or identical epitopes are recognized for both proteins based on the high level of amino acid sequence identity. In addition, ATEM1 protein is readily detected in extracts from seeds homozygous for the atem6-1 allele. Because the coding region of the gene is unchanged and the mRNA is appropriately spliced, protein translated from that mRNA should display all the characteristics of wild-type protein, including stability. We hypothesize, therefore, that the decrease in protein levels is due, at least in part, to altered stability of the chimeric mRNA. This is supported by the presence of significantly lower levels of the ATEM6 transcript in the homozygous mutant. In addition, if polyadenylation is poor or inefficient, there could be a significant effect on the process of ribosome recycling (Munroe and Jacobson, 1990; Gallie, 1991; Gallie and Tanguay, 1994) that might further hinder the efficiency of translation for the mRNA species that are present.
The severe reduction or elimination of ATEM6 protein production is manifested in homozygous mutant plants as a distinct phenotype during late seed maturation. At a time in development when seeds from wild-type plants are still fully green and hydrated along the length of the silique, mutant seeds at the distal end of siliques display premature drying. In addition, other aspects of the maturation process appear to be accelerated as evidenced by the accumulation of pigments in those same seeds. Display of the phenotype associated with elimination of this single group 1 LEA protein corresponds to the normal developmental time for the onset of ATEM6 expression, and beyond 14 daf the remaining mutant seeds mature rapidly and relatively synchronously. This phenotype is not present in plants heterozygous for the atem6-1 mutation, suggesting that the level of ATEM6 protein produced from a single wild-type allele is sufficient for normal seed development. Complementation of the atem6-1 mutation using a 3,002-bp ATEM6 allele restored the wild-type phenotype of the homozygous mutant seed. This demonstrates that the presence of ATEM6 is necessary to maintain hydration throughout the length of the silique during the later stages of seed development.
Premature desiccation was not observed in all mutant siliques and occasionally there was early drying of a few wild-type siliques. However, the frequency of premature drying in atem6-1 plants was significantly higher and much more severe than the number observed in wild-type plants (Supplemental Table I). This suggests that the phenotype associated with the atem6-1 mutation is variably expressed. Statistical analysis (lsd-ANOVA) was used to determine whether the phenotype of a population of wild-type plants was statistically different from that of a population of atem6-1 mutants and demonstrated that the phenotypes of the two populations are not the same (P value < 0.001; Supplemental Table II). lsd-ANOVA analysis was also used to compare the phenotypes of wild-type, atem6-1 mutant, and atem6-1 complement populations. The results indicated that the phenotype of the complementation plants is not different from the wild type (P value < 0.001) and that the atem6-1 mutant phenotype is different from both the wild-type and complement plants (P value < 0.001; Supplemental Table II.). A three-way contingency table was used to determine whether any of the silique phenotypes could be due to chance or whether they were based on plant type (wild type, mutant, or complement). The results indicated that the phenotypes could only be due to plant type (P = 0.001; Supplemental Table III).
Several studies have demonstrated that rearrangements, deletions, and inversions may be a relatively frequent occurrence at sites of T-DNA integration (Castle and Meinke, 1994; Tax and Vernon, 2001). Data from the plasmid rescue of the atem6-1 T-DNA revealed deletions associated with the LB and RB of the T-DNA, but no deletions of ATEM6 genomic sequences. In an effort to verify that the chromosome was otherwise largely intact (aside from the insertion itself), PCR-based markers were used to map the large arm of chromosome 2. Each of the three markers mapped to the correct chromosome and in the correct order when compared to ATEM6 and to each other. These data indicate that there were no major aberrations associated with the atem6-1 insertion, and the phenotype of the mutant may be attributed solely to the atem6-1 mutation.
Based on our observations, ATEM6 protein expression is necessary for normal seed development. While characterization of the atem6-1 mutant plants at this level does not allow us to directly address some of the functions proposed for group 1 LEA proteins (e.g. the water replacement hypothesis), the phenotype suggests that expression of ATEM6 protein may be required to buffer the rate of dehydration during the latter stages of seed maturation. This is consistent with the very hydrophilic nature of group 1 LEA proteins, yet does not preclude involvement in other mechanisms. Our hypothesis is also consistent with initial display of the phenotype in seeds that are most distal from the remainder of the plant because those would be the first to experience lower water potentials. Interestingly, this mutation is not lethal and mutant seeds are indistinguishable from wild type visually and readily germinate under laboratory conditions. We are currently exploring the possibility that expression of the other group 1 LEA, ATEM1, displays an altered level or domain of expression in mutant seeds that might in some way compensate for loss of ATEM6 expression. An alternative, although not mutually exclusive, explanation is that the near-optimal conditions used for plant propagation in the laboratory setting do not reveal a decrease in fitness that might be apparent in mutant plants grown in the wild. The exact mechanisms by which the group 1 LEA proteins contribute to dehydration/desiccation tolerance remain to be fully elucidated.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis (Arabidopsis thaliana) plants in this work are of the Columbia-0 ecotype. The putative T-DNA insertions in ATEM6 were identified in the Syngenta Arabidopsis Insertion Library (Sessions et al., 2002). Plants were grown on soil (Fafard 3B potting media) in pots with a 16-h photoperiod at 24°C ± 2°C.
Plasmid Rescue Experiments
Leaf material was collected, ground under liquid nitrogen, and chromosomal DNA was extracted as described previously (Krysan et al., 1996). Chromosomal DNA (10 μg) was digested overnight with either EcoRI or NcoI under standard conditions. Digests were extracted with phenol:CHCl3:isoamyl alcohol (25:24:1; v/v) and precipitated with ethanol. The resulting fragments were then subjected to dilute ligation in a 10-mL total volume to promote circularization and transformed into Escherichia coli strain DH5αMCR, using ampicillin at 100 μg/mL for selection. Plasmid DNA was sequenced using the Big Dye version 3.0 kit according to the manufacturer's instructions (Applied Biosystems). Plasmids rescued using EcoRI digestion were sequenced using a T7 promoter primer and the QRB2 primer (Table III). Plasmids rescued using NcoI digestion were sequenced with a T3 promoter primer and the QLB2 or AJM-LB1 primer (Table III).
Mapping of Chromosome 2
Homozygous atem6-1 plants (Columbia ecotype) were crossed to wild-type Landsberg erecta. Heterozygous F1 plants were allowed to self-pollinate, generating a segregating F2 population. F2 plants were grown on soil and leaf tissue was collected from 100 individuals for genomic DNA extraction.
Leaves were removed and immediately minced in a microcentrifuge tube with a plastic pestle. Each sample was vortexed with 200 μL of extraction buffer (100 mm Tris-HCl, pH 9.5; 1 m KCl; 10 mm EDTA) and incubated at 65°C for 30 min. Samples were centrifuged at 13,000 rpm at room temperature for 10 min. The supernatant was precipitated with 500 μL 100% ethanol, placed at −20°C for 30 min, and then centrifuged at 13,000 rpm at room temperature for 15 min. DNA pellets were washed with 70% ethanol, centrifuged at 13,000 rpm for 2 min at room temperature, and resuspended in 10 mm Tris HCl, pH 8.0, 0.1 mm EDTA (Tris-EDTA).
All PCR reactions were run using the following components: 10 mm Tris-HCl, pH 8.3, 50 mm KCl, 2 mm MgCl2, 0.2 mm dNTPs, 12 pmol each primer (see below), and 2 units of Taq polymerase (all primer sequences are listed in Table III). The NGA168 marker (NGA168-F and NGA168-R primers) was amplified with the following program: 94°C, 5 min for one cycle; 94°C, 1 min, 49°C, 1 min, 72°C, 1 min for 10 cycles; 94°C, 1 min, 51°C, 1 min, 72°C, 1 min for 15 cycles; and 72°C, 7 min. The ML and Thy1 markers, the ATEM6 allele, and the atem6-1 allele were amplified using the following program with varying annealing temperatures (Ta): 94°C, 5 min for one cycle; 94°C, 1 min, Ta, 1 min, 72°C, 1 min for 35 cycles; and 72°C, 7 min. The ML-F and ML-R primers used a Ta of 51°C, the Thy1-F and Thy1-R primers used a Ta of 50°C, the ATEM6 allele (ATEM6 gene-specific 2 and ATEM6 gene-specific 3 primers) used a Ta of 55°C, and the atem6-1 allele (ATEM6 gene-specific 2 and AJM-LB1 primers) used a Ta of 50°C. All PCR products were size fractionated on a 0.7% agarose gel.
Southern-Blot Analysis
Plasmid and genomic DNAs were subjected to restriction endonuclease digestion and size fractionated on 0.7% agarose gels at 15 to 40 V overnight. The DNA was then blotted onto a Hybond N+ membrane using a downward alkaline transfer (Chomczynski, 1992) and cross-linked to the membrane using UV light (Strata-Linker; Stratagene). Gene-specific 3-′UTR or T-DNA-specific (pBS) probe fragments were labeled with [α-32P]dATP using a random octamer reaction (Feinberg and Vogelstein, 1983, 1984). Membranes were hybridized at 65°C overnight in a buffer containing 4× sodium chloride/sodium phosphate EDTA (SSPE), 6% polyethylene glycol (PEG) 8000, 0.5% SDS, 2× Denhardt's solution, and 10 mg/mL sheared salmon sperm DNA. After hybridization, the membranes were quickly rinsed with a buffer of 5× SSPE, 0.1% SDS followed by a quick rinse in a 2× SSPE, 0.1% SDS solution (all posthybridization washing solutions were prewarmed to 65°C). The membranes were then washed twice for 20 min each with 2× SSPE, 0.1% SDS, 1× SSPE, 0.1% SDS, and finally with 0.1× SSPE, 0.1% SDS for 10 to 15 min. Membranes were exposed to a phosphorimaging screen for 4 h to overnight, depending on signal intensity, and imaged on a Typhoon 9400 variable mode imager (Amersham Biosciences).
RT-PCR
Ten seeds were pulverized at room temperature in a sterile conical microcentrifuge tube with a plastic pestle cleaned with 95% ethanol. RNA was extracted from the seeds according to the protocol described (Chomczynski and Sacchi, 1987). The dried RNA pellets were resuspended in 10 μL of the annealing mixture (10 mm Tris-HCl, pH 8.3, 50 mm KCl, 1.5 mm MgCl2, 25 pmol tailed oligo(dT) primer [Table III]) for the RT reaction. This mixture was heated to 65°C to 70°C for 5 to 10 min and allowed to slow cool to 30°C to 35°C. Once cooled, 10 μL of extension mix (10 mm Tris-HCl, pH 8.3, 50 mm KCl, 1.5 mm MgCl2, 2 mm dithiothreitol, 1 mm dNTPs, 5 units AMV-RT [Promega]) were added and the reaction was incubated at 42°C for 60 min. Finally, 2 μL of the RT reaction were subjected to PCR (10 mm Tris-HCl, pH 8.3, 50 mm KCl, 2 mm MgCl2, 0.2 mm dNTPs, 25 pmol each primer [ATEM6 RT-PCR forward and reverse; Table III], 2 units of Taq polymerase). The PCR cycle was 94°C, 5 min for one cycle; 94°C, 1 min, 50°C, 1 min, 72°C, 1 min for 25 to 30 cycles; and 72°C, 7 min.
RNA Gel-Blot Analysis
RNA was extracted as described (Vicient and Delseny, 1999) from roughly 100 μL of seed ground into a fine powder under liquid nitrogen. RNA samples (10 μg) were separated on a 1.2% agarose-formaldehyde gel. The RNA was capillary blotted onto Hybond N+ membrane using diethyl pyrocarbonate-treated 10× SSC as the transfer buffer and cross-linked to the membrane using UV light (Strata-Linker; Stratagene). An ATEM6 gene-specific 5′-UTR probe fragment was labeled with [α-32P]dATP using a random octamer reaction (Feinberg and Vogelstein, 1983, 1984). Membranes were hybridized at 65°C overnight in a modified Church and Gilbert buffer containing 0.5 m NaPO4, pH 7.2, 7% SDS, 10 mm EDTA, pH 8.0. After hybridization, the membranes were quickly rinsed with a buffer of 5× SSPE, 0.1% SDS followed by a quick rinse in a 2× SSPE, 0.1% SDS solution. All posthybridization washing solutions were prewarmed to 65°C. The membranes were then washed twice for 20 min each with 2× SSPE, 0.1% SDS, 1× SSPE, 0.1% SDS, and finally with 0.1× SSPE, 0.1% SDS for 10 to 15 min. Membranes were exposed to a phosphorimaging screen (4 h to overnight), imaged on a Typhoon 9400 variable mode imager, and images analyzed using ImageQuant software (Amersham Biosciences).
Immunoblot Analysis
Protein was extracted from a single silique minced in a microcentrifuge tube containing 200 μL of an extraction buffer containing 10 mm Tris-HCl, pH 8.3, and 10 mm NaCl. The tubes were vortexed well, then centrifuged for 2 min at 10K rpm at 4°C. The supernatant was removed from the seed tissue and put into a new microcentrifuge tube and centrifuged two or three times until no solid material remained at the bottom of the tube. Extracts were then frozen at −20°C and put directly into the centrifuge at maximum speed at 4°C for 30 min. The supernatant was transferred to a new tube and heated to 80°C for 20 min followed by centrifugation at maximum speed at 4°C for 30 min. This supernatant was transferred to a fresh tube and precipitated with 25% TCA. Samples were incubated on ice for 5 min, then centrifuged at maximum speed at 4°C for 5 min. The supernatant was removed and the protein pellets were washed with 100% acetone and centrifuged for another 5 min at 4°C. The acetone was removed, the pellets dried slightly, and resuspended directly in gel-loading buffer. Protein extracts were resolved on a 15% Tris-Gly gel and electrotransferred to an Immobilon-P PVDF membrane (Millipore) at 100 V for 1 h at 4°C. Blots were processed using a primary polyclonal rabbit anti-wheat Em antibody, a secondary alkaline phosphatase-conjugated goat anti-rabbit antibody. Chemiluminescent detection was performed using ready-to-use CSPD (Roche Diagnostics).
atem6-1 Complementation Construct Preparation
A 3,002-bp ATEM6 genomic clone was PCR amplified using the ATEM6-F and ATEM6-R primers (Table III) and cloned into the SmaI site of pBS (Stratagene). The genomic clone and pBI101 vector were digested using BamHI and EcoRI, run on a 0.7% agarose gel, and the desired fragments isolated. Insert and vector were ligated and transformed into E. coli strain DH5αMCR, using kanamycin at 20 μg/mL for selection. The resulting clones were confirmed using a HindIII/EcoRI digest. Correct clones were transformed into Agrobacterium (Agrobacterium tumefaciens; strain LBA4404) using simple heat shock (Chen et al., 1994).
Agrobacterium Transformation of Arabidopsis
Pots containing 10 to 20 homozygous atem6-1 mutant plants were grown until the first inflorescence meristems reached a length of at least 10 cm. Agrobacterium cultures were grown as described (Clough and Bent, 1998). Yeast extract peptone media (50 mL) was supplemented with 20 μg/mL kanamycin sulfate for cultures grown overnight. Cells were pelleted at 5,500g for 20 min at 22°C and resuspended to an OD600 of 0.8 to 1.0 in a solution of 5% Suc containing 0.05% Silwet L-77 (Osi Specialties). Flowers and buds of each racime were inoculated with the resuspended culture. Plants were covered and kept in the dark overnight, then returned to normal growth conditions the next day. Plants were allowed to grow until maturity and dry seed was harvested.
Selection of Transformed Seeds
Seeds were surface sterilized with 70% ethanol for 1 min, 10% bleach, containing 0.1% Triton X-100 for 10 min and rinsed with sterile distilled water three times. Germination media consisted of 0.5× Murashige and Skoog media (one packet of Murashige and Skoog minimal salts [no. M6899; Sigma] supplemented with 1% Suc, 0.8 mg/L thiamine, 0.5 mg/mL pyridoxine, 0.5 mg/mL nicotinic acid, and 50 mg/L myoinositol) and 0.8% Phytagar was supplemented with 50 μg/mL kanamycin sulfate and 125 μg/mL carbenicillin. Sterile seeds were mixed with 0.5× Murashige and Skoog Phytagar germination media and poured onto selection plates. To prevent drying or contamination while allowing gas exchange, plates were wrapped with latex-free medical bandaging tape.
Plates were cold treated at 4°C for 3 d, then moved into the growth chamber. Seeds remained on the selection media for 7 d until green seedlings with first true leaves were identified. These seedlings were placed on 0.5× Murashige and Skoog nonselective media for 7 d, then moved onto soil.
PCR was used to confirm that the selected plants contained both the atem6-1 allele and the wild-type ATEM6 complement. All PCRs were done using 10 mm Tris-HCl, pH 8.3, 50 mm KCl, 2 mm MgCl2, 0.2 mm dNTPs, 12 pmol each primer (see below), and 2 units of Taq polymerase. The following program (with variable Ta) was used for all PCRs: 94°C, 5 min for one cycle; 94°C, 1 min, Ta, 1 min, 72°C, 1 min for 30 cycles; and 72°C, 7 min. The ATEM6 gene-specific 2 and AJM LB-1 primers (Ta = 50°C) were used to amplify the atem6-1 allele. The ATEM6 gene-specific 2 and ATEM6 gene-specific 3 primers (Ta = 55°C) were used to amplify the wild-type ATEM6 allele. Primers specific to the kanamycin NptII gene (Ta = 50°C) present within the pBI101 T-DNA were also used to verify the presence of the complementation T-DNA. All primer sequences are listed in Table III.
Phenotype Analysis
The phenotypes of seeds within the silique were determined by dissecting the silique from base or pedicel to tip using Dumont number 4 and 5 forceps. The silique was held in place at the pedicel while the forceps were used to dissect longitudinally along the natural line of dehiscence. One-half of each valve covering the seeds was removed to expose the seeds while keeping them in situ. Images of the seeds were taken with a Nikon SMZ1500 dissecting scope using Q-Capture software (version 2.68.6; Q-Imaging). A score of wild type was assigned if the seeds were desiccating synchronously along the length of the silique. A score of mutant was assigned if the first one to five seeds in the silique were mature and desiccated, while the remaining seeds were green. The numerical values from the phenotype study (percent wild type-looking siliques) were converted to a normal distribution using an arcsine-squared conversion. Statistical analysis of phenotype was done using Systat software (version 10; Systat).
Supplementary Material
Acknowledgments
We thank Julie M. Long for technical assistance and Julia Frugoli for critical reading of the manuscript. We also thank Torrey Mesa Research Institute/Syngenta for providing the putative Arabidopsis T-DNA insertion lines.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture (USDA) Cooperative State Research, Education and Extension Service (CSREES; grant no. 2002–35318–12627 to W.R.M.); the CSREES/USDA (project no. SC–1700200 to W.R.M.); the Howard Hughes Medical Institute SC-LIFE Undergraduate Research Program; and the Clemson University Calhoun Honors College, Technical Contribution Number 5061 of the Clemson University Experiment Station.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: William R. Marcotte Jr. (marcotw@clemson.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.072967.
References
- Baker J, Steele C, Dure L III (1988) Sequence and characterization of 6 Lea proteins and their genes from cotton. Plant Mol Biol 11: 277–291 [DOI] [PubMed] [Google Scholar]
- Bartels D, Singh M, Salamini F (1988) Onset of desiccation tolerance during development of the barley embryo. Planta 175: 485–492 [DOI] [PubMed] [Google Scholar]
- Berge SK, Bartholomew DM, Quatrano RS (1989) Control of the expression of wheat embryo genes by abscisic acid. In RL Goldberg, ed, Molecular Basis of Plant Development. Alan R. Liss, New York, pp 193–201
- Bies N, Aspart L, Carles C, Gallois P, Delseny M (1998) Accumulation and degradation of Em proteins in Arabidopsis thaliana: evidence for post-transcriptional controls. J Exp Bot 49: 1925–1933 [Google Scholar]
- Bostock RM, Quatrano RS (1992) Regulation of Em gene expression in rice. Interaction between osmotic stress and abscisic acid. Plant Physiol 98: 1356–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW (1993) Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet 241: 504–514 [DOI] [PubMed] [Google Scholar]
- Castle LA, Meinke DW (1994) A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell 6: 25–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Nelson RS, Sherwood JL (1994) Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16: 664–670 [PubMed] [Google Scholar]
- Chomczynski P (1992) One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal Biochem 201: 134–139 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cuming A (1999) LEA proteins. In P Shewry, R Casey, eds, Seed Proteins. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 753–780
- Dure L III (1993) Structural motifs in lea proteins. Curr Topics Plant Physiol 10: 91–103 [Google Scholar]
- Dure L III, Crouch M, Harada J, Ho THD, Mundy J, Quatrano R, Thomas T, Sung ZR (1989) Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol 12: 475–486 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1984) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Addendum. Anal Biochem 137: 266–267 [DOI] [PubMed] [Google Scholar]
- Feldmann K (1992) T-DNA insertion mutagenesis in Arabidopsis: seed infection/transformation. In C Koncz, N-H Chua, J Schell, eds, Methods in Arabidopsis Research. World Scientific Publishing, Singapore, pp 274–289
- Gallie DR (1991) The cap and poly(A) tail function synergistically to regulate messenger-RNA translational efficiency. Genes Dev 5: 2108–2116 [DOI] [PubMed] [Google Scholar]
- Gallie DR, Tanguay R (1994) Poly(A) binds to initiation factors and increases cap-dependent translation in vitro. J Biol Chem 269: 17166–17173 [PubMed] [Google Scholar]
- Gaubier P, Raynal M, Hull G, Huestis GM, Grellet F, Arenas C, Pages M, Delseny M (1993) Two different Em-like genes are expressed in Arabidopsis thaliana seeds during maturation. Mol Gen Genet 238: 409–418 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR (1996) Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA 93: 8145–8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Meinke DW (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 16: 21–31 [DOI] [PubMed] [Google Scholar]
- McCubbin WD, Kay CM, Lane BG (1985) Hydrodynamic and optical properties of the wheat-germ Em protein. Can J Biochem Cell Biol 63: 803–811 [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris PC, Kumar A, Bowles DJ, Cuming AC (1990) Osmotic stress and abscisic acid induce expression of the wheat Em genes. Eur J Biochem 190: 625–630 [DOI] [PubMed] [Google Scholar]
- Munroe D, Jacobson A (1990) mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol 10: 3441–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russouw PS, Farrant J, Brandt W, Lindsey GG (1997) The most prevalent protein in a heat-treated extract of pea (Pisum sativum) embryos is an LEA group I protein; its conformation is not affected by exposure to high temperature. Seed Sci Res 7: 117–123 [Google Scholar]
- Russouw PS, Farrant J, Brandt W, Maeder D, Lindsey GG (1995) Isolation and characterization of a heat-soluble protein from pea (Pisum sativum) embryos. Seed Sci Res 5: 137–144 [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulages JL, Kim K, Walters C, Cushman JC (2002) Temperature-induced extended helix/random coil transitions in a group 1 late embryogenesis-abundant protein from soybean. Plant Physiol 128: 822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy RA, Espelund M, Saeboe-Larssen S, Hollung K, Helliesen E, Jakobsen KS (1995) Evolution of the group 1 late embryogenesis abundant (Lea) genes: analysis of the Lea B19 gene family in barley. Plant Mol Biol 28: 1039–1054 [DOI] [PubMed] [Google Scholar]
- Tax FE, Vernon DM (2001) T-DNA-associated duplication/translocations in Arabidopsis. Implications for mutant analysis and functional genomics. Plant Physiol 126: 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EW, Lane BG (1980) Relation of protein synthesis in imbibing wheat embryos to the cell-free translational capacities of bulk mRNA from dry and imbibing embryos. J Biol Chem 255: 5965–5970 [PubMed] [Google Scholar]
- Vicient C, Hull G, Guilleminot J, Devic M, Delseny M (2000) Differential expression of the Arabidopsis genes coding for Em-like proteins. J Exp Bot 51: 1211–1220 [PubMed] [Google Scholar]
- Vicient CM, Delseny M (1999) Isolation of total RNA from Arabidopsis thaliana seeds. Anal Biochem 268: 412–413 [DOI] [PubMed] [Google Scholar]
- Welin BV, Olson A, Nylander M, Palva ET (1994) Characterization and differential expression of Dhn/Lea/Rab-like genes during cold-acclimation and drought stress in Arabidopsis thaliana. Plant Mol Biol 26: 131–144 [DOI] [PubMed] [Google Scholar]
- Wilhelm KS, Thomashow MF (1993) Arabidopsis thaliana Cor15b, an apparent homolog of Cor15a, is strongly responsive to cold and ABA, but not drought. Plant Mol Biol 23: 1073–1077 [DOI] [PubMed] [Google Scholar]
- Williamson JD, Quatrano RS (1988) ABA-regulation of two classes of embryo-specific sequences in mature wheat embryos. Plant Physiol 86: 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ (2003) LEAping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics 4: 52–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.