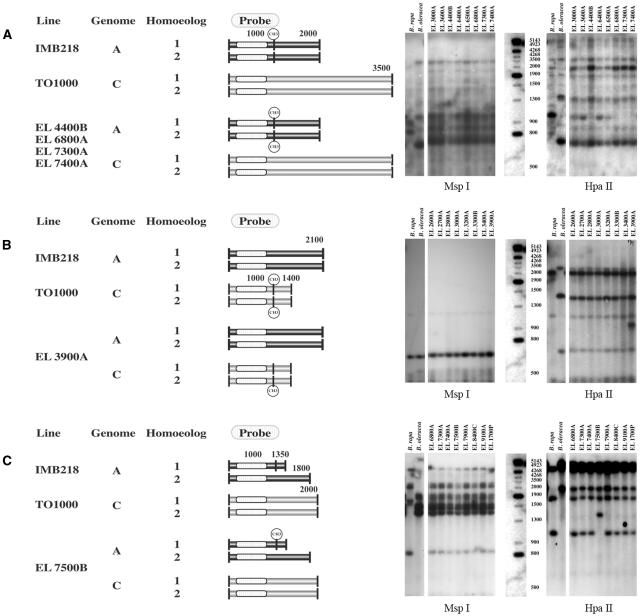
Figure 4.
Changes in the restriction fragment pattern of HpaII digests between parental and polyploid genomes. Observed fragment patterns on Southern blots of HpaII restriction digests from B. oleracea, B. rapa, and pooled DNAs from the progeny of individual S0 polyploids (right). All S0 progeny DNAs had parental MspI restriction fragments. Models of the putative methylation changes that generated the observed hybridization pattern are depicted to the left. The two horizontal rectangles associated with a single genome represent homoeologous loci. Each vertical line represents a HpaII recognition site, the sequence CCGG. A solid vertical line represents a site that does not have cytosine C5 methylation within the CpG dinculeotide sequence. A vertical line with a methyl group represents a CpG methylated site. A, Loss of a B. rapa-specific fragment (1 kb) is not accompanied by the gain of a novel fragment, or the novel fragment comigrates with preexisting fragments and cannot be unambiguously scored. Here, hybridization of HpaII-restricted DNA with pX144 shows a high-Mr fragment (2 kb) has a greater hybridization intensity among lines with a missing 1-kb fragment (EL4400B, EL6800A, EL7300A, and EL7400A) than among lines without the missing fragment. This pattern suggests that de novo methylation occurred within the polyploid A genome. All parental fragments are shown. B, Gain of a novel fragment within allopolyploids as seen by hybridization of HpaII-restricted DNA with pW157. This pattern may suggest that demethylation occurred at one of two or more homoeologous loci. All parental fragments are shown. C, Loss of a parental fragment and gain of a larger, novel fragment within the allopolyploid as seen by hybridization of HpaII-restricted DNA with pW125. De novo cytosine methylation within the S0 parent at a site unmethylated within the diploid B. rapa progenitor explains this pattern. All parental fragments are shown.