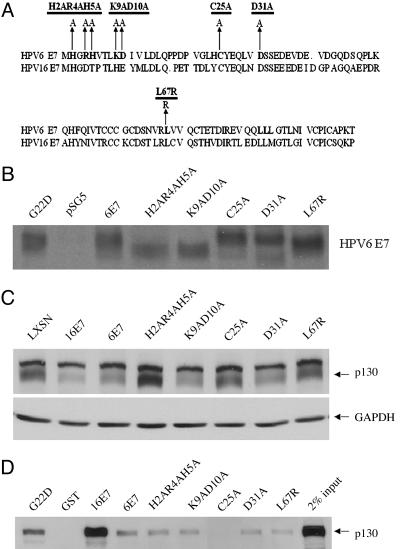
Fig. 4.
Construction of HPV-6 E7 mutants and their characterization with respect to p130 binding and degradation. (A) Map of HPV-6 E7 mutants. (B) Stability of mutants. COS-7 cells were transfected with pSG5, pSG5(6E7), pSG5(H2AR4AH5A), pSG5(K9AD10A), pSG5(C25A), pSG5(D31A), and pSG5(L67R). Forty-eight hours after transfection, the cells were labeled with 500 μCi of 35S-labeled methionine and cysteine and lysed, and HPV-6 E7 was immunoprecipitated by a polyclonal antibody (anti-6E7). The immunoprecipitates were separated on 15% SDS/PAGE gel and exposed to x-ray film. (C) Amino acids required for destabilization of p130. HFKs were infected with LXSN, L(16E7)SN, L(6E7)SN, or retroviruses encoding the HPV-6 E7 mutants. Whole-cell lysates from transduced cells grown in C-K-SFM were analyzed by Western blot with antibodies to p130. The average ± SD of p130 in three independent experiments, corrected for GAPDH and relative to LXSN, was 0.33 ± 0.06 (16E7), 0.35 ± 0.06 (6E7), 1.03 ± 0.05 (H2AR4AH5A), 0.43 ± 0.06 (K9AD10A), 0.79 ± 0.15 (C25A), 0.37 ± 0.07 (D31A), and 0.89 ± 0.32 (L67R). (D) E7 amino acids required for binding to p130. GST pull-downs were conducted as in Fig. 1 with GST, GST-16E7, GST-6E7H2AR4AH5A, GST-6E7K9AD10A, GST-6E7C25A, GST-6E7D31A, and GST-6E7L67R, and the immunoblots were probed with antibodies to p130. Ponceau red staining indicated equal amounts of GST and GST fusion proteins in the precipitate (data not shown). The average ± SD of bound p130 in three independent experiments, corrected for Ponceau red and relative to GST6E7, was 6.86 ± 2.40 (16E7), 1.23 ± 0.76 (H2AR4AH5A), 0.51 ± 0.18 (K9AD10A), 0.16 ± 0.09 (C25A), 1.12 ± 0.12 (D31A), and 0.71 ± 0.05 (L67R).