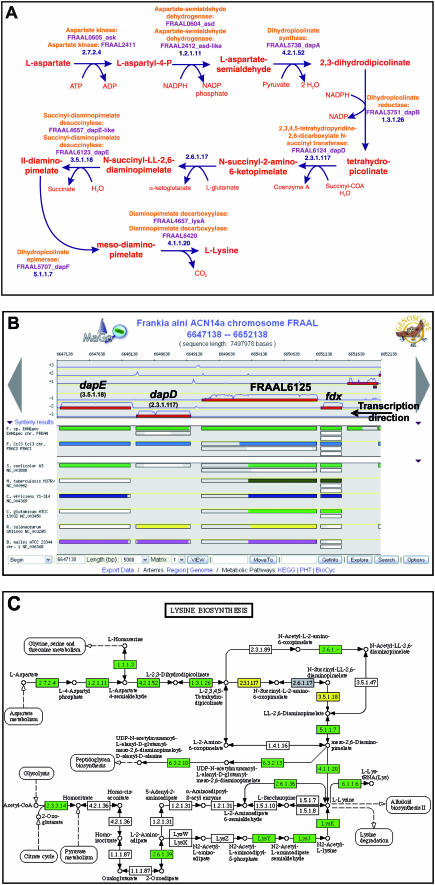
Figure 4.
Lysine biosynthesis in F.alni genome through the MaGe interfaces. Three screenshots showing lysine biosynthesis in F.alni. The FrankiaCyc Pathway/Genome DataBase (PGDB) is available through MaGe via a BioCyc web server (A). In addition, the user can obtain KEGG maps by comparison with E.coli (C). Yellow rectangles symbolize enzymes encoded by genes in the selected MaGe region (B) while green rectangles represent enzymes encoded by genes localized elsewhere in the studied genome. Gray boxes correspond to known enzymes in E.coli that are not present in the genome under study. Lastly, white boxes are enzymatic activities missing in both organisms. The BioCyc pathway selection algorithm reports only one possible pathway for lysine biosynthesis (A) in F.alni. The reported pathway apparently lacks the gene(s) encoding the succinyldiaminopimelate amino transferase activity (EC number 2.6.1.17). The lysine biosynthesis map from KEGG (C) also reports the lack of succinyldiaminopimelate amino transferase activity which has been detected in E.coli. Furthermore, genomic context exploration of the genes involved in this pathway, via the MaGe genome browser (B), reveals that the gene FRAAL6125 is co-localized with the characterized dapE and dapD genes. FRAAL6125 is a good candidate for dapC, a gene coding the missing activity and experimentally described in other species.