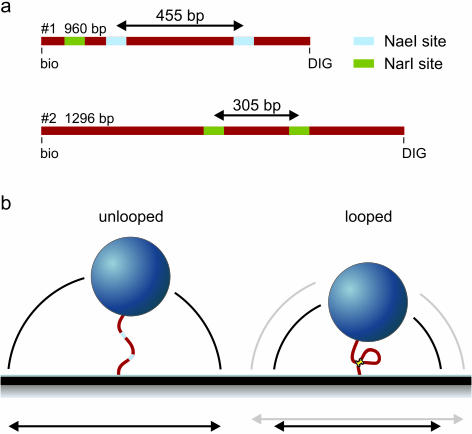
Figure 2.
(a) DNA templates used in the experiments. Template #1 is 960 bp in length and has two NaeI recognition sites (and one NarI site). Template #2 is slightly longer, 1296 bp and harbors two NarI sites. In the looped state template #1 is 505 bp and template #2 991 bp in length. (b) Schematic representation of the experiment. Small beads are tethered with the DNA molecule in question to the glass slide. By tracking the x- and y-positions of the bead the magnitude of the Brownian motion is monitored, which is a measure for the tether length. Upon DNA loop formation by a restriction enzyme the Brownian motion of the bead suddenly decreases. This allows following DNA looping kinetics in real-time.