Abstract
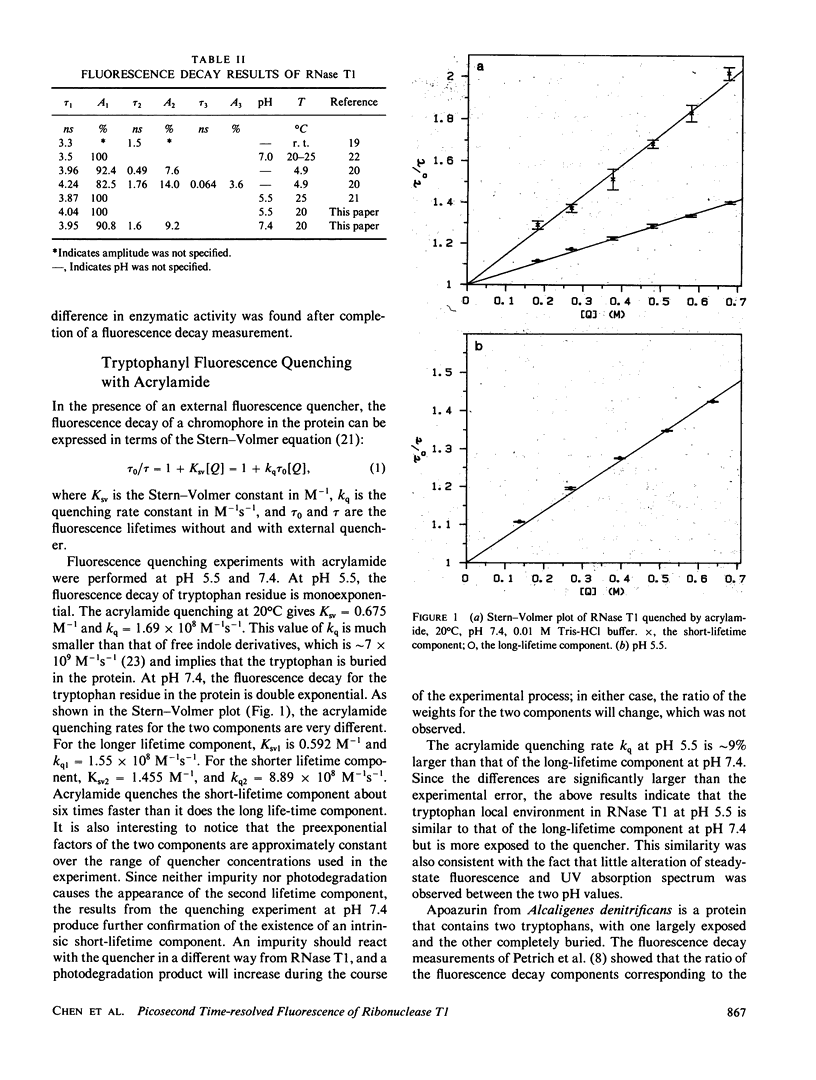
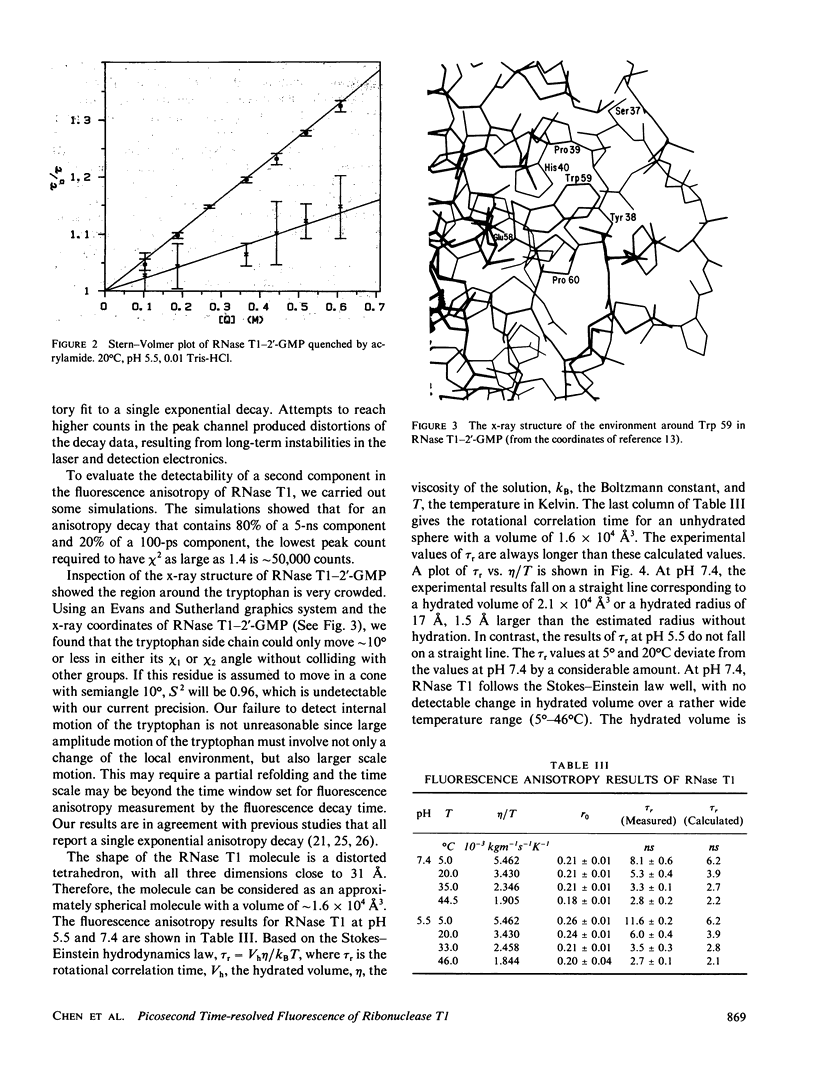
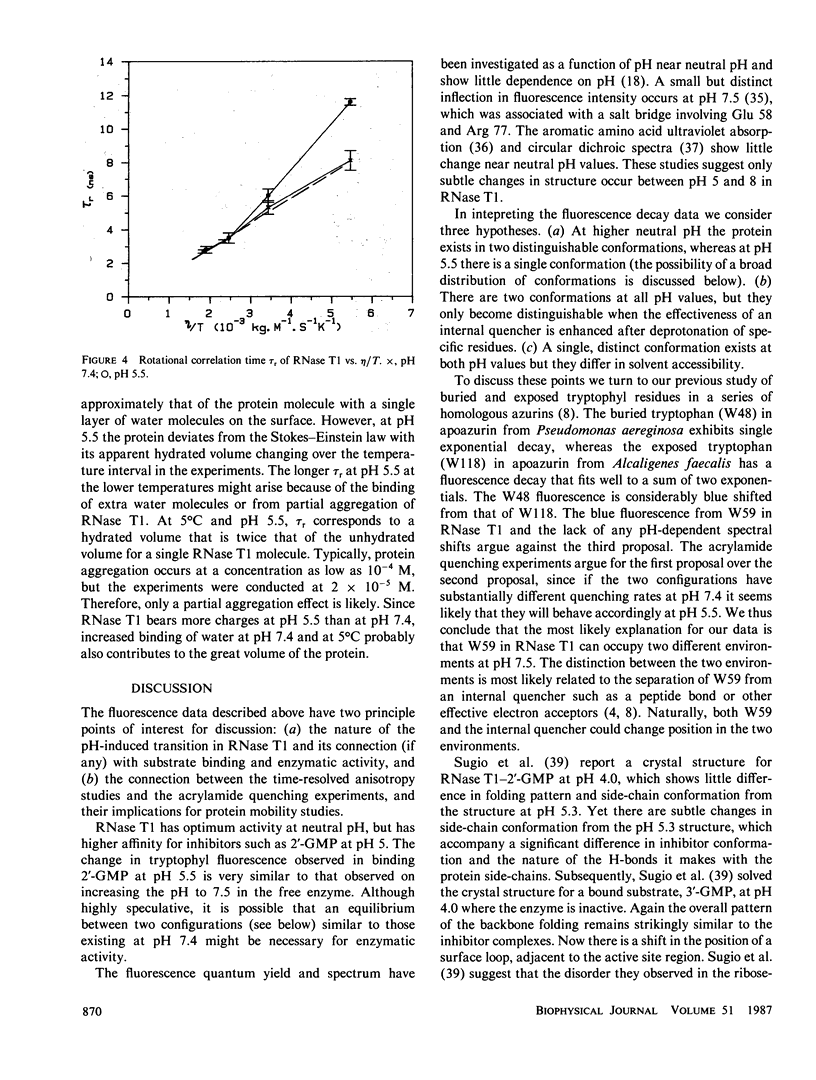
The tryptophyl fluorescence of ribonuclease T1 decays monoexponentially at pH 5.5, tau = 4.04 ns but on increasing pH, a second short-lived component of 1.5 ns appears with a midpoint between pH 6.5 and 7.0. Both components have the same fluorescence spectrum. Acrylamide quenches both fluorescence components, and the short-lived component is quenched fivefold faster than the predominant long component. Binding of the substrate analogue 2'-guanylic acid at pH 5.5 quenches the fluorescence by 20% and introduces a second decay component, tau = 1.16 ns. Acrylamide quenches both tryptophyl decay components, with similar quenching rates. The fluorescence anisotropy decay of ribonuclease T1 was consistent with a molecule the size of ribonuclease T1 surrounded by a single layer of water at pH 7.4, even though the anisotropy decay at pH 5.5 deviated from Stokes-Einstein behavior. The fluorescence data were interpreted with a model where the tryptophyl residue exists in two conformations, remaining in a hydrophobic pocket. The acrylamide quenching is interpreted with electron transfer theory and suggests that one conformer has the nearest atom approximately 3 A from the protein surface, and the other, approximately 2 A.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arata Y., Kimura S., Matsuo H., Narita K. Proton and phosphorus nuclear magnetic resonance studies of ribonuclease T1. Biochemistry. 1979 Jan 9;18(1):18–24. doi: 10.1021/bi00568a003. [DOI] [PubMed] [Google Scholar]
- Beechem J. M., Brand L. Time-resolved fluorescence of proteins. Annu Rev Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- Calhoun D. B., Vanderkooi J. M., Englander S. W. Penetration of small molecules into proteins studied by quenching of phosphorescence and fluorescence. Biochemistry. 1983 Mar 29;22(7):1533–1539. doi: 10.1021/bi00276a003. [DOI] [PubMed] [Google Scholar]
- Chang M. C., Cross A. J., Fleming G. R. Internal dynamics and overall motion of lysozyme studied by fluorescence depolarization of the eosin lysozyme complex. J Biomol Struct Dyn. 1983 Oct;1(1):299–318. doi: 10.1080/07391102.1983.10507441. [DOI] [PubMed] [Google Scholar]
- Cross A. J., Fleming G. R. Analysis of time-resolved fluorescence anisotropy decays. Biophys J. 1984 Jul;46(1):45–56. doi: 10.1016/S0006-3495(84)83997-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftink M. R., Ghiron C. A. Dynamics of a protein matrix revealed by fluorescence quenching. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3290–3294. doi: 10.1073/pnas.72.9.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftink M. R., Ghiron C. A. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry. 1976 Feb 10;15(3):672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- Eftink M. Quenching-resolved emission anisotropy studies with single and multitryptophan-containing proteins. Biophys J. 1983 Sep;43(3):323–334. doi: 10.1016/S0006-3495(83)84356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinvald A., Steinberg I. Z. The fluorescence decay of tryptophan residues in native and denatured proteins. Biochim Biophys Acta. 1976 Apr 14;427(2):663–678. doi: 10.1016/0005-2795(76)90210-5. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Saenger W. Specific protein-nucleic acid recognition in ribonuclease T1-2'-guanylic acid complex: an X-ray study. Nature. 1982 Sep 2;299(5878):27–31. doi: 10.1038/299027a0. [DOI] [PubMed] [Google Scholar]
- Hershberger M. V., Maki A. H., Galley W. C. Phosphorescence and optically detected magnetic resonance studies of a class of anomalous tryptophan residues in globular proteins. Biochemistry. 1980 May 13;19(10):2204–2209. doi: 10.1021/bi00551a032. [DOI] [PubMed] [Google Scholar]
- Inagaki F., Kawano Y., Shimada I., Takahashi K., Miyazawa T. Nuclear magnetic resonance study on the microenvironments of histidine residues of ribonuclease T1 and carboxymethylated ribonuclease T1. J Biochem. 1981 Apr;89(4):1185–1195. [PubMed] [Google Scholar]
- James D. R., Demmer D. R., Steer R. P., Verrall R. E. Fluorescence lifetime quenching and anisotropy studies of ribonuclease T1. Biochemistry. 1985 Sep 24;24(20):5517–5526. doi: 10.1021/bi00341a036. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R., Maliwal B. P., Cherek H., Balter A. Rotational freedom of tryptophan residues in proteins and peptides. Biochemistry. 1983 Apr 12;22(8):1741–1752. doi: 10.1021/bi00277a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterman H. L., Walz F. G., Jr Subsite interactions and ribonuclease T1 catalysis: kinetic studies with APGpC and ApGpU. Biochemistry. 1979 May 15;18(10):1984–1988. doi: 10.1021/bi00577a021. [DOI] [PubMed] [Google Scholar]
- Pongs O. Influences of pH and substrate analogs on ribonuclease T1 fluorescence. Biochemistry. 1970 May 26;9(11):2316–2322. doi: 10.1021/bi00813a015. [DOI] [PubMed] [Google Scholar]
- Ross J. B., Schmidt C. J., Brand L. Time-resolved fluorescence of the two tryptophans in horse liver alcohol dehydrogenase. Biochemistry. 1981 Jul 21;20(15):4369–4377. doi: 10.1021/bi00518a021. [DOI] [PubMed] [Google Scholar]
- Sander C., Ts'o P. O. Circular dichroism studies on the conformation and interaction of T 1 ribonuclease. Biochemistry. 1971 May 25;10(11):1953–1966. doi: 10.1021/bi00787a001. [DOI] [PubMed] [Google Scholar]
- Sato S., Egami F. On the interaction of ribonuclease T-1 and guanosine 2'-phosphate and related compounds. Biochem Z. 1965 Aug 19;342(4):437–448. [PubMed] [Google Scholar]
- Sugio S., Oka K., Ohishi H., Tomita K., Saenger W. Three-dimensional structure of the ribonuclease T1 X 3'-guanylic acid complex at 2.6 A resolution. FEBS Lett. 1985 Apr 8;183(1):115–118. doi: 10.1016/0014-5793(85)80966-2. [DOI] [PubMed] [Google Scholar]
- TAKAHASHI K. The structure and function of ribonuclease T1. II. Further purification and amino acid composition of ribonuclease T1. J Biochem. 1962 Feb;51:95–108. doi: 10.1093/oxfordjournals.jbchem.a127515. [DOI] [PubMed] [Google Scholar]
- Takahashi K. A revision and confirmation of the amino acid sequence of ribonuclease T1. J Biochem. 1985 Sep;98(3):815–817. doi: 10.1093/oxfordjournals.jbchem.a135339. [DOI] [PubMed] [Google Scholar]
- Takahashi K. The amino acid sequence of ribonuclease T-1. J Biol Chem. 1965 Oct;240(10):4117–4119. [PubMed] [Google Scholar]
- Takahashi K. The structure and function of ribonuclease T1. 18. Gel filtration studies on the interaction of ribonuclease T1 with substrate analogs. J Biochem. 1972 Dec;72(6):1469–1481. doi: 10.1093/oxfordjournals.jbchem.a130039. [DOI] [PubMed] [Google Scholar]
- Turoverov K. K., Kuznetsova I. M., Zaitsev V. N. The environment of the tryptophan residue in Pseudomonas aeruginosa azurin and its fluorescence properties. Biophys Chem. 1985 Nov;23(1-2):79–89. doi: 10.1016/0301-4622(85)80066-1. [DOI] [PubMed] [Google Scholar]
- Walz F. G., Jr, Hooverman L. L. Interaction of guanine ligands with ribonuclease T1. Biochemistry. 1973 Nov 20;12(24):4846–4851. doi: 10.1021/bi00748a006. [DOI] [PubMed] [Google Scholar]
- Walz F. G., Jr Studies on the nature of guanine nucleotide binding with ribonuclease T1. Biochemistry. 1977 Dec 13;16(25):5509–5515. doi: 10.1021/bi00644a018. [DOI] [PubMed] [Google Scholar]
- Walz F. G., Jr, Terenna B. Subsite interactions of ribonuclease T1: binding studies of dimeric substrate analogues. Biochemistry. 1976 Jun 29;15(13):2837–2842. doi: 10.1021/bi00658a021. [DOI] [PubMed] [Google Scholar]


