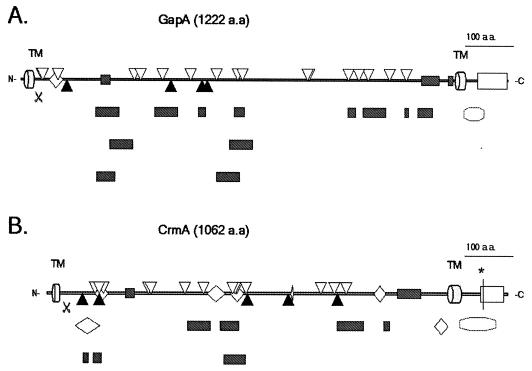
FIG. 5.
Putative binding and interactive domains of M. gallisepticum GapA (A) and CrmA (B). [ ], transmembrane region (TM); [
], transmembrane region (TM); [ ], putative signal peptidase cleavage site; [
], putative signal peptidase cleavage site; [ ], domains shared by carbohydrate binding proteins (determined by performing a BLOCKS database search [26]); [▿], heparin sulfate binding-like domains (determined by performing a FastA search [58] with data from references 27, 48, 64, 65, and 70); [▴], lectin-like recognition domains (determined by performing a Patternmatch search with data from references 49 and 67); [◊], other domains shared by some adhesion molecules (determined by performing a RPS-BLAST search on the Conserved Domain database [2]); [□], interaction domains shared proteins involved in the signal transduction-MARCKS family (determined by performing a BLOCKS database search [26]); [], histone H1, HMG14/HMG17 signature (GapA), H2B and H5 signature, HMG14/HMG17 family, DNA polymerase family X, and AlgR homology (CrmA) (determined by performing a BLOCKS database search [26]). [∗], ATP/GTP-binding site motif A (P-loop) (determined by performing a PROSITE motif search [11]).
], domains shared by carbohydrate binding proteins (determined by performing a BLOCKS database search [26]); [▿], heparin sulfate binding-like domains (determined by performing a FastA search [58] with data from references 27, 48, 64, 65, and 70); [▴], lectin-like recognition domains (determined by performing a Patternmatch search with data from references 49 and 67); [◊], other domains shared by some adhesion molecules (determined by performing a RPS-BLAST search on the Conserved Domain database [2]); [□], interaction domains shared proteins involved in the signal transduction-MARCKS family (determined by performing a BLOCKS database search [26]); [], histone H1, HMG14/HMG17 signature (GapA), H2B and H5 signature, HMG14/HMG17 family, DNA polymerase family X, and AlgR homology (CrmA) (determined by performing a BLOCKS database search [26]). [∗], ATP/GTP-binding site motif A (P-loop) (determined by performing a PROSITE motif search [11]).