Abstract
Human T cell leukemia virus type I (HTLV-I) causes adult T cell leukemia (ATL) in 2–5% of carriers after a long latent period. An HTLV-I encoded protein, Tax, induces proliferation and inhibits apoptosis, resulting in clonal proliferation of infected cells. However, tax gene expression in ATL cells is disrupted by several mechanisms, including genetic changes in the tax gene and DNA methylation/deletion of the 5′ long terminal repeat (LTR). Because Tax is the major target of cytotoxic T-lymphocytes in vivo, loss of Tax expression should enable ATL cells to escape the host immune system. The 5′ LTR of HTLV-I is frequently hypermethylated or deleted in ATL cells, whereas the 3′ LTR remains unmethylated and intact, suggesting the involvement of the 3′ LTR in leukemogenesis. Here we show that a gene encoded by the minus strand of the HTLV-I proviral genome, HTLV-I basic leucine zipper factor (HBZ), is transcribed from 3′-LTR in all ATL cells. Suppression of HBZ gene transcription by short interfering RNA inhibits proliferation of ATL cells. In addition, HBZ gene expression promotes proliferation of a human T cell line. Analyses of T cell lines transfected with mutated HBZ genes showed that HBZ promotes T cell proliferation in its RNA form, whereas HBZ protein suppresses Tax-mediated viral transcription through the 5′ LTR. Thus, the single HBZ gene has bimodal functions in two different molecular forms. The growth-promoting activity of HBZ RNA likely plays an important role in oncogenesis by HTLV-I.
Keywords: oncogenesis, retrovirus, bimodal function
Human retroviruses use their genomes very efficiently because of their limited genome size. Their accessory genes elaborately control replication of genome copy (1). The HIV vigorously replicates to yield progeny virus, whereas human T cell leukemia virus type I (HTLV-I) increases the number of infected cells by the activity of accessory genes (2, 3). HTLV-I was the first human retrovirus associated with human disease (4, 5). After transmission of HTLV-I, 2–5% of carriers are likely to develop adult T cell leukemia (ATL) after a long latent period (6). HTLV-I belongs to the δ-retrovirus group, which includes bovine leukemia virus and simian T-cell leukemia virus. In contrast to HIV, HTLV-I is transmitted in a cell-to-cell fashion requiring transfer of infected cells (7). To facilitate transmission, HTLV-I increases the number of infected cells through the activity of accessory genes, which are encoded by the pX region located between env and the 3′ long terminal repeat (LTR). These genes include tax, rex, p30, p12, p13, and HTLV-I basic leucine zipper factor (HBZ) (3, 8). Among them, tax is thought to play a central role in increasing the number of infected cells. Tax activates transcriptional pathways, including nuclear factor κ-B, cAMP response element-binding protein, activator protein-1, and serum responsive factor (2, 3). In addition, Tax can functionally inactivate p53 (9), resulting in inhibition of apoptosis. Thus, Tax promotes proliferation and suppresses apoptosis of infected cells, leading to clonal proliferation (10–12). As a consequence, HTLV-I causes ATL, a fatal neoplastic disease of CD4-positive T-lymphocytes.
Despite its critical role in proliferation of infected cells, Tax expression in ATL cells is disrupted by several mechanisms, including genetic changes in the tax gene (13), DNA methylation (14, 15), or deletion of the 5′ LTR (16). Because Tax is the major target of cytotoxic T-lymphocytes (17), Tax-expressing cells are rapidly eliminated in vivo. Therefore, loss of Tax expression could enable ATL cells to evade the host immune system. On the other hand, the role of HTLV-I-encoded viral genes has not yet been determined in ATL cells that lack Tax expression. In ATL cells, the HTLV-I 3′ LTR remains unmethylated and intact (18), whereas the 5′ LTR is frequently hypermethylated or deleted. Based on these observations, we hypothesized that promoter/enhancer activity of the HTLV-I 3′ LTR was essential for proliferation and survival of ATL cells. Transcription from the minus strand of HTLV-I has been reported (19), and the HBZ was subsequently found to inhibit Tax-mediated transactivation of viral gene transcription from the 5′ LTR by heterodimerizing with either cAMP response element-binding protein 2, c-Jun or JunB (20, 21).
In this study, we found that the HBZ gene was expressed in all ATL cells and that suppression of HBZ transcription by short interfering RNA (siRNA) decreased ATL cell proliferation. Mutant analyses showed that the HBZ gene promoted proliferation of T cells in its RNA form, whereas HBZ protein inhibited Tax-mediated transactivation through the HTLV-I LTR. These findings suggest that HBZ plays an important role in oncogenesis by HTLV-I.
Results
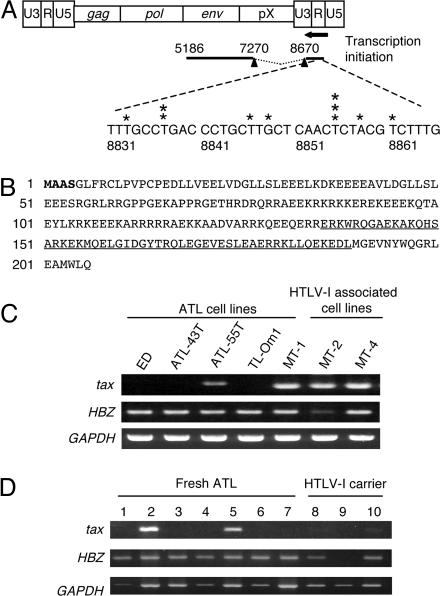
The Spliced Form of HBZ Is Expressed in All ATL Cells. We first determined the transcription start site of HBZ by using 5′ RACE (Fig. 1A). Contrary to a previous report (20), the HBZ gene was spliced and transcriptional start sites were identified in the R and U5 region of the 3′ LTR. The first 4 amino acids of the predicted HBZ protein differed from the previously reported sequence (Fig. 1B). The 3′ end of the transcript was also determined by 3′ RACE (Fig. 1 A). A polyadenylation signal was found in 3′ untranslated region of HBZ. The spliced HBZ gene does not overlap with the tax-encoding region, indicating that an antisense RNA to tax is not generated. We next analyzed HBZ transcription in ATL cell lines, fresh ATL cells, and peripheral blood mononuclear cells from HTLV-I carriers by RT-PCR. In three ATL cell lines (ED, ATL-43T, and TL-Om1), tax gene transcription was silenced, whereas HBZ transcription was detected in all cell lines (Fig. 1C). HBZ was transcribed in all seven fresh ATL cell samples, whereas tax gene transcription was observed in two cases (Fig. 1D). Furthermore, HBZ gene transcription was detected in two of three carriers. Although genetic changes (nonsense mutations, insertions, and deletions) in tax have been reported in refs. 13 and 15, HBZ sequences did not contain disruptive genetic changes in any cell line or in fresh ATL samples (17 cases), except for polymorphisms (Table 1, which is published as supporting information on the PNAS web site).
Fig. 1.
HBZ gene expression in ATL cells. (A)5′ RACE was performed by using total RNA from the ATL cell line ATL-55T. The schema represents the HTLV-I provirus and spliced HBZ mRNA. Asterisks show transcription initiation sites identified by 5′ RACE. The 3′ end of the transcript (5186) was identified by 3′ RACE, and polyadenylation signal was found upstream (5206–5211) of this transcript. Nucleic acids are numbered with reference to ATK-1 according to Seiki et al. (22). (B) Hypothetical amino acid sequence derived from spliced HBZ. Amino acids different from the previously reported HBZ are shown in bold type. The basic leucine zipper domain is underlined. (C) Expression of tax and HBZ genes in ATL and HTLV-I-immortalized cell lines analyzed by RT-PCR. (D) Expression of tax and HBZ genes in fresh ATL cells and peripheral blood mononuclear cells from HTLV-I carriers. Lanes: 1–7, fresh ATL cases; 8–10, peripheral blood mononuclear cells from HTLV-I carriers.
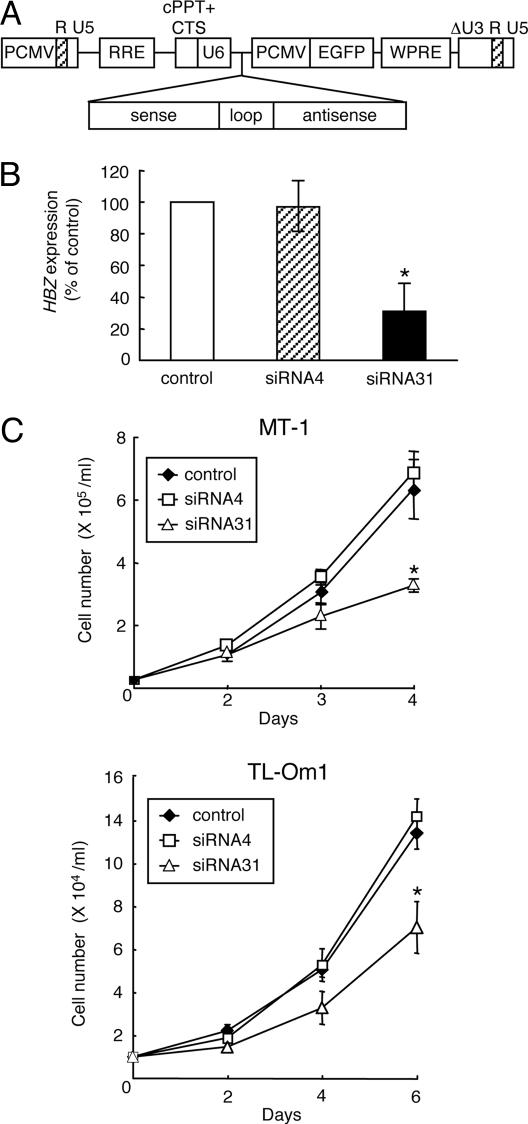
Inhibition of HBZ by siRNA Suppresses Growth of ATL Cells. Although inhibition of Tax-mediated viral gene transcription has been reported as a function of HBZ (20), HBZ was expressed in ATL cells lacking tax gene transcription, suggesting that HBZ has other roles. To clarify the role of HBZ in ATL cells, we suppressed HBZ gene expression by using siRNAs. Recombinant lentiviruses transcribing short hairpin RNAs against HBZ under control of the U6 promoter (Fig. 2A) were generated and transduced to tax-expressing MT-1 and tax-nonexpressing TL-Om1 cells. In MT-1 cells, siRNA31 suppressed HBZ gene mRNA expression, whereas siRNA4 did not (Fig. 2B). Growth of both cell lines was suppressed by siRNA31 (Fig. 2C) along with HBZ transcription (Fig. 6, which is published as supporting information on the PNAS web site). Apoptosis was not induced by siRNA31. These findings indicate that HBZ gene expression is required for ATL cell proliferation, regardless of tax gene expression.
Fig. 2.
Knockdown of HBZ gene expression inhibits cell growth of ATL cells. (A) Schematic diagram of the short hairpin RNA-expressing lentiviral vector. PCMV, immediate early cytomegalovirus promoter; RRE, Rev-responsive element; cPPT, central polypurine tract; CTS, central termination sequence; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element. (B) siRNA31 decreases HBZ gene transcription, whereas siRNA4 does not (Supporting Methods, which is published as supporting information on the PNAS web site). Efficiencies of lentivirus vector transfection, which were determined by EGFP expression, were 99.3% and 93.0% for MT-1 and TL-Om1, respectively. HBZ transcripts in siRNA transfectants of MT-1 are quantified by a densitometer and shown as percentages compared with a mock transfectant. Values are means ± SD from three independent experiments. (C) Transfection of lentivirus vector expressing siRNA31 suppresses the growth of MT-1 and TL-Om1. The transfectants of siRNAs were harvested at day 7 after transfection, and seeded into a 96-well plate at 5 × 103 cells per well. Cell numbers of each transfectants were counted in triplicate by Trypan blue dye exclusion method. Values are means ± SD. *, P < 0.05.
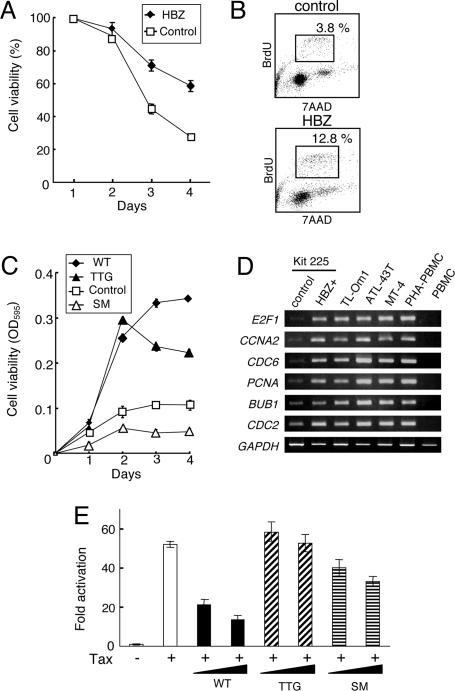
To analyze HBZ function, a vector expressing HBZ was transfected into Kit 225 cells, an IL-2-dependent human T cell line derived from T cell chronic lymphocytic leukemia cells (23), and stable transformants were selected in the presence of G418. HBZ-transfected Kit 225 cells showed prolonged survival after removal of IL-2 (Fig. 3A) compared with control cells, and HBZ-transduced Kit 225 cells showed an increase in the number of cells in S phase after depletion of IL-2 (Fig. 3B). In addition, HBZ-transfected Kit 225 cells showed enhanced responsiveness to IL-2 (Fig. 3C, WT). To identify HBZ target genes in Kit 225 cells, transcriptional profiles of HBZ-transfected and control cells after withdrawal of IL-2 (48 h) were analyzed by an oligonucleotide microarray. We identified 687 genes up-regulated (by >2-fold) and 719 genes down-regulated (by >2-fold) in HBZ-transfected cells (Tables 2 and 3, which are published as supporting information on the PNAS web site). Expression of selected candidate genes was confirmed by RT-PCR (Fig. 3D), which indicated that transcription of the E2F1 gene and its targets, CCNA2, CDC6, PCNA, CDC2, and BUB1, were up-regulated in HBZ-transfected Kit 225 cells. We speculated that up-regulation of E2F1 is involved in enhanced proliferation mediated by HBZ.
Fig. 3.
Functional analyses of the HBZ gene. (A) HBZ gene expression prolongs survival of transfected Kit 225 cells after withdrawal of IL-2. Cell viabilities are measured with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay. Values are means ± SD. (B) Cell cycle analyses of HBZ-expressing Kit 225 cells, and Kit 225 cells transfected with a control vector. Cells in S phase were identified by BrdUrd incorporation and staining with 7AAD after withdrawal of IL-2. (C) Effects of wild-type (WT) and mutant forms of HBZ on proliferation of IL-2-stimulated Kit 225 cells. A suboptimal level of IL-2 (2.5 units/ml) was added after removal of IL-2 (48 h), and cell viability was measured by an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay. The first ATG of HBZ is mutated to TTG, which blocks synthesis of HBZ protein. All codons in the HBZ gene are replaced with silent mutations (SM). The sequence of SM HBZ has been shown in Fig. 7. (D) Semiquantitative RT-PCR analysis for differentially expressed genes in HBZ-expressing Kit 225 cells compared with control cells. (E) Effects of HBZ and its mutants on Tax-mediated transactivation through the HTLV-I LTR. Lusiferase reporter (WT-Luc) and Tax expressing vector were transfected into Jurkat cells with or without vectors expressing wild-type (WT) or mutated HBZ (TTG or SM) (0.3 or 1 μg). Fold activation were calculated compared with basal luciferase activity of WT-Luc. Results are means ± SD in triplicate.
To confirm whether HBZ protein promotes cell proliferation, we generated vectors expressing two HBZ gene mutants. Mutation that the first ATG of HBZ is replaced by TTG blocks synthesis of HBZ protein (TTG HBZ). Surprisingly, expression of TTG HBZ still promoted proliferation (Fig. 3C), suggesting that HBZ functions to promote cell proliferation as an RNA. To confirm this hypothesis, we replaced all coding regions in the HBZ gene with silent mutations (SM HBZ) (Fig. 7, which is published as supporting information on the PNAS web site). Such mutations completely altered the secondary structure of the RNA, but the SM HBZ gene still produced intact HBZ protein. SM HBZ gene expression did not promote cell proliferation (Fig. 3C), indicating that the growth-promoting effect of HBZ depended on RNA structure.
Because an inhibitory effect of HBZ on Tax-mediated transactivation through the 5′ LTR has been reported (20), we analyzed this function by using mutant forms of HBZ. As shown in Fig. 3E, wild-type HBZ suppressed Tax-mediated transactivation of transcription from the HTLV-I 5′ LTR. Although the SM HBZ gene also suppressed transactivation, TTG HBZ completely lacked this activity. Taken together, these findings suggest that HBZ promotes T cell proliferation in the form of RNA, whereas HBZ protein inhibits Tax-mediated transactivation through the 5′ LTR.
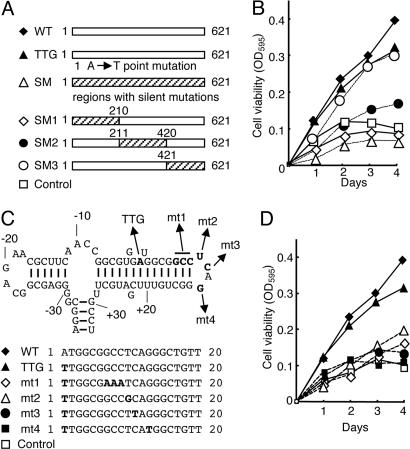
To further analyze the growth-promoting activity of HBZ RNA, we constructed vectors expressing a series of mutated HBZ genes (Fig. 4 A and C). When HBZ genes with silent mutations in different regions were analyzed, we found that mutations in the first 210 bp of HBZ (SM1) abolished growth-promoting activity (Fig. 4B). In addition, silent mutations in SM2 partially decreased growth-promoting activity of HBZ. When we compared the predicted secondary RNA structures of the native HBZ gene and the form of HBZ expressed in the pME18Sneo vector, the first stem-loop of HBZ RNA was present in both cases (Fig. 4C). Although the TTG HBZ gene retained strong growth-promoting activity, TTG HBZ genes with mutations in the first stem-loop structure showed reduced or no activity (Fig. 4 C and D, mutants 1–4), indicating that it is essential for growth-promoting activity.
Fig. 4.
Functional analyses of mutated HBZ genes on proliferation of T cells. (A) Schemas of mutant HBZ genes. (B) The effects of mutated HBZ genes on cell growth were measured by assays. The hatched area represents the region containing silent mutations. Numbers indicate the nucleotide positions in the HBZ coding sequence. (C) Predicted stem-loop structure in HBZ mRNA of the native HBZ gene (from –35 to +33). Nucleotides are numbered from the first ATG (A: +1) of the HBZ gene. Structural predictions were analyzed by using mfold (24). Mutated sequences of each vector are shown in bold type. (D) The results of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assays with these vectors are shown.
Microarray analyses of Kit 225 cells expressing wild-type and TTG HBZ genes identified transcriptional changes mediated by HBZ RNA, showing that RNA was responsible for up-regulation of E2F1 (Table 2). Taken together, we conclude that the first stem-loop structure is important for growth-promoting activity of HBZ. To study the possibility that microRNA is responsible for growth-promoting activity, we performed Northern blot analysis by using oligonucleotides from the region containing the first stem-loop structure as probes. However, microRNA was not detected by Northern blot analysis (data not shown).
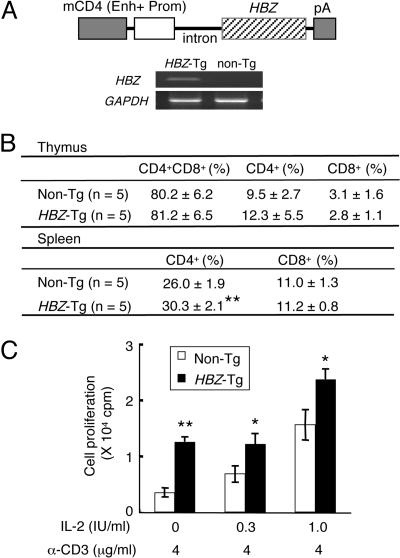
In Vivo Effect of HBZ Gene Expression in Transgenic Mice. To analyze the role of HBZ in vivo, we generated transgenic mice expressing HBZ under the control of the mouse CD4 promoter/enhancer, which induces specific transgene expression in CD4-positive cells (Fig. 5A) (25). HBZ expression in transgenic mice was confirmed in CD4-positive splenocytes by RT-PCR (Fig. 5A). Thymocyte subpopulations in transgenic mice did not differ from those observed in nontransgenic littermates. However, the percentage of CD4-positive T lymphocytes increased in splenocytes of transgenic mice (P < 0.01) (Fig. 5B). In addition, proliferation induced by cross-linking with an immobilized anti-CD3 antibody was augmented in thymocytes of transgenic mice (Fig. 5C). These data indicate that the HBZ gene promotes proliferation of CD4-positive T lymphocytes in vivo.
Fig. 5.
Generation and analyses of HBZ-transgenic (Tg) mice. (A) Schematic representation of the HBZ transgene. The promoter (Prom) and enhancer (Enh) of the mouse CD4 (mCD4) gene were ligated to HBZ cDNA (including the 5′ UTR) plus the polyadenylation signal sequence of SV40. Expression of HBZ transcripts was detected by RT-PCR in purified CD4+ splenocytes from HBZ-Tg mice. (B) T cell subsets in HBZ-Tg mice. Values are means ± SD from five transgenic mice. **, P < 0.01. (C) Proliferative responses of thymocytes from HBZ-Tg mice to IL-2 and/or stimulation with an anti-CD3 antibody. Proliferative responses were measured by 3H-thymidine incorporation. 3H-thymidine uptake of thymocytes without anti-CD3 antibody and IL-2 was 5.8 ± 4.6 cpm for control mice and 21.0 ± 16.0 cpm for HBZ transgenic mice. Values are means ± SD in triplicate. *, P < 0.05; **, P < 0.01.
Discussion
HTLV-I infection causes a fatal neoplastic disease, ATL, after a long latent period. Because transfection of retroviral vectors expressing tax can immortalize T lymphocytes in vitro (26), it has been suggested that tax plays a central role in oncogenesis among HTLV-I accessory genes. Tax has pleiotropic actions by interacting with cellular proteins, which promotes clonal proliferation of infected cells (2, 3). However, the role of the tax gene remains an enigma in ATL cells, because tax gene transcripts cannot be detected in ≈60% of ATL cases (12). However, the 3′ LTR and HBZ sequences are conserved in all ATL cells regardless of deletion or hypermethylation of the 5′ LTR or of genetic changes in the tax gene, suggesting that an intact 3′ LTR and HBZ gene are critical for oncogenesis. In this study, we showed that the HBZ gene encoded by the minus strand of the HTLV-I provirus is transcribed in all ATL cells. HBZ has been reported to suppress Tax-mediated transactivation of viral gene transcription through the 5′ LTR. In addition to this function, we showed that HBZ promotes proliferation of T lymphocytes in vitro and in vivo. These findings indicate that HBZ plays a critical role in oncogenesis mediated by HTLV-I, even in late stages of oncogenesis when tax is not expressed.
In oncogenic DNA viruses, oncoproteins, such as the SV40 T antigen and human papillomavirus E7 protein, target retinoblastoma (Rb) protein and inactivate its function (27). Loss of Rb function is associated with deregulation of E2F transcription factors (28). E2F1, a critical regulator of cell cycle progression, plays a pivotal role in the G1-to-S-phase transition by transactivating specific target genes (29). Furthermore, overexpression of E2F1 has been reported to be associated with oncogenesis (30). As shown in this study, HBZ RNA increased E2F1 gene transcription, which is likely associated with HBZ-induced proliferation. Although Tax was also reported to induce E2F1 expression (31), E2F1 is overexpressed in ATL cell lines lacking Tax expression, indicating that HBZ may be responsible for up-regulating E2F1 expression.
Tax promotes cell growth and inhibits apoptosis of infected cells. However, because Tax is the major target of cytotoxic T lymphocytes, its expression is also disadvantageous to the survival of infected cells (17, 32). Therefore, HTLV-I encodes several other viral genes, such as rex, p30, and HBZ, to suppress Tax production by different mechanisms (20, 21, 33, 34). Because such suppression has a negative effect on growth of infected cells, the HBZ gene is likely to support proliferation of infected cells in addition to its activity in suppressing tax gene transcription. Because growth-promoting activity of HBZ could be a critical factor in maintaining a leukemic state, HBZ should be a therapeutic target.
Recently, microRNAs have been demonstrated to play important roles in regulating gene expression (35). Virus-encoded microRNAs have been also identified (36, 37), and some function in viral replication. In HIV-1, microRNAs have been demonstrated to function in viral transcription (38, 39). In HTLV-I, when env RNA is transcribed with the HBZ gene, such a long double-stranded RNA could be precursor for Dicer processing, resulting in formation of viral siRNAs. Such siRNA might function to suppress viral gene expression in HTLV-I. It is noteworthy that HBZ transcription is suppressed in MT-2 cells (Fig. 1C). In MT-2 cells, env-tax fusion gene is abundantly transcribed (15), which might generate siRNA and suppress HBZ expression.
Epstein–Barr virus (EBV)-encoded nonpolyadenylated RNA (also known as EBV-encoded small RNA or EBER) is known to function in oncogenesis by activating transcription of genes such as insulin-like growth factor 1 (40) or interleukin 9 (41). Although these functional RNAs do not encode polypeptides, HBZ can function as both RNA and protein. Because of their limited genome size (8,506 bp for HTLV-I), complex retroviruses have evolved to use RNA splicing to express required genes. In addition to such a mechanism, HTLV-I not only expresses the HBZ gene on the minus strand but it also utilizes this gene as protein and RNA. Bimodal functions of viral gene may represent a previously uncharacterized strategy to regulate viral replication and proliferation of infected cells.
In conclusion, we showed that spliced HBZ gene was transcribed in all ATL cells. The HBZ gene promotes proliferation of ATL cells in the RNA form, whereas HBZ protein suppresses Tax-mediated viral transcription through the 5′ LTR. Although the role of tax gene remains undetermined in ATL cells, this study sheds light on the role of HBZ gene in oncogenesis.
Materials and Methods
Cells. Two HTLV-I immortalized lines (MT-2, and MT-4) and five ATL cell lines (ED, ATL-43T, ATL-55T, TL-Om1, and MT-1) were used in this study (15). The IL-2-dependent human T cell line Kit 225 was maintained in RPMI medium 1640 supplemented with 10% FBS and recombinant IL-2 (85 units/ml). Approval for this study was obtained from the institutional review board of Kyoto University. Informed consent was obtained from blood donors and patients according to the Declaration of Helsinki. To construct vectors encoding wild-type (WT) and mutant forms of HBZ, the coding sequence (621 bp) was amplified from cDNA of TL-Om1 cells and subcloned into the pME18Sneo vector (42). Vectors were transfected into Kit 225 cells by using Nucleofector (Amaxa Biosystems, Cologne, Germany). Briefly, cells were suspended in 100 μl of Cell Line Nucleofector Solution T and then nucleofected with vectors (5 μg) by using program T-16 of the Nucleofector device (Amaxa Biosystems). Stable transfectants were selected in G418 (600 μg/ml).
Rapid Amplification of cDNA 5′and 3′ Ends (RACE). To determine the 5′ and 3′ ends of transcripts, RACE was performed by using the SMART RACE cDNA amplification kit (BD Biosciences Clontech) according to manufacturer's instructions (42). First-strand cDNAs were synthesized from 1 μg of total RNA of ATL-55T, ATL-43T, or MT-1 cells by reverse transcriptase and used for 5′ RACE PCR. For nested amplifications, primers specific for the HBZ gene (5′-CCTCTTTCTCCCGCTCTTTTTT TCGC-3′ and 5′-CATGACACAGGCAAGCATCGAAACA-3′) were used. For nested 3′ RACE amplifications, primers specific for the HBZ gene (5′-CTAGGTTAGGGCAGGGGGGCTGTAGGGC-3′ and 5′-GGGTCCACGAACAAACTGGCTGGGCAGG-3′) were used. PCR products were cloned into vectors, and the nucleotide sequences were determined.
Synthesis of cDNA and Semiquantitative RT-PCR. Spliced HBZ, tax, and GAPDH transcripts were quantified by using RT-PCR. The primers used were as follows: HBZ gene: 5′-TAAACTTACCTAGACGGCGG-3′ (sense), 5′-CTGCCGATCACGATGCGTTT-3′ (antisense); tax gene: 5′-CCGGCGCTGCTCTCATCCCGGT-3′ (sense) and 5′-GGCCGAACATAGTCCCCCAGAG-3′ (antisense). PCR was performed in a PC-808 (Astec) under the following conditions: HBZ, 2 min at 95°C, followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 57.5°C, and 30 seconds at 72°C; tax, 2 min at 95°C, followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 61°C, and 30 seconds at 72°C. The intensity of PCR-amplified band was quantified by using ATTO densitograph 4.0 (Atto Instruments, Tokyo). Semiquantitative RT-PCR was performed to confirm microarray results by using primers (Table 4, which is published as supporting information on the PNAS web site).
Lentiviral Vector Construction and Transfection of Recombinant Lentivirus. We modified pCSII-EF-MCS (43) for delivery of anti-HBZ short hairpin RNAs, and recombinant lentivirus was produced as described in the Supporting Methods. The titer of concentrated virus stocks was measured on 293 T cells based on their EGFP expression. Cells were then transfected with concentrated vector stocks at a multiplicity of infection of 10–25 in the presence of polybrene (4 μg/ml, Sigma). Cells were harvested 7 days later, and EGFP expression of transfected cells was analyzed with an EPICS XL Flow Cytometer (Beckman Coulter). When >90% of transfected cells expressed EGFP, cell growth and HBZ gene expression were analyzed.
Microarray Analysis. Total RNAs were isolated from Kit 225 cells, which were transfected with a vector expressing the HBZ gene, the TTG HBZ gene (the first ATG of HBZ gene was replaced by TTG), or with a control vector by using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Total RNAs were then purified by RNeasy (Qiagen). Oligonucleotide microarray analyses (CodeLink, Human 20K I bioarrays, Amersham Pharmacia Biosciences) were performed by Kurabo Industries (Osaka) with the authorization of Amersham Pharmacia Biosciences. A 2.0-fold increase or decrease was considered significant, based on the manufacturer's recommendation.
Luciferase Assay. Jurkat cells were grown in RPMI medium 1640 containing 10% FBS. On day 1, cells were seeded into 6-well plates at 4 × 105 cells per well. After 24 h, cells were transfected with 1 μg per well of luciferase reporter plasmid (WT-Luc) (44), 40 ng per well of pRL-TK (Promega), and pCG-Tax (1 μg) (45), and/or an HBZ expression plasmid (0.3 or 1 μg), and/or blank expression vector (to normalize the DNA dose) mixed with Transfectin (Bio-Rad). After 48 h, cells were collected and luciferase activities were measured by using a Dual Luciferase Reporter Assay Kit (Promega).
Generation of Transgenic Mice. HBZ cDNA was cloned into the SalI site of the H/M/T-CD4 vector, which was designed for restricted expression of a transgene in CD4+ cells (25). The purified fragment containing the HBZ transgene was microinjected into C57BL/6J F1 fertilized eggs. Transgenic founders were screened for integration of transgenes by PCR and mated with C57BL/6J mice to generate transgenic progeny. All animals used in this study were maintained and handled according to protocols approved by Kyoto University.
Cell Proliferation Assay. Proliferation assays of murine cells were carried out in RPMI 1640 medium supplemented with 10% FBS and 2-mercaptoethanol (50 μM). Thymocytes (1 × 106 cells per ml) were stimulated by an immobilized anti-CD3 antibody (4 μg/ml) with or without recombinant IL-2 in flat-bottomed 96-well plates. Thymocyte proliferation was measured by 3H-thymidine uptake after 3 days of incubation. Cell viabilities were assessed by measuring 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide dye absorbance (42).
Flow Cytometric Analysis. Cell cycles of HBZ-transfected and control Kit 225 cells were analyzed after removal of IL-2 (48 h) by using BrdU Flow Kits (Becton Dickinson Pharmingen) according to the manufacturer's instructions. To analyze CD4 and CD8 expression in HBZ transgenic mice, cells (5 × 105) were reacted with monoclonal antibodies against murine CD4 (FITC-labeled, Immunotech) and CD8 (phycoerythrin-labeled, Immunotech), according to the manufacturer's instructions, and analyzed with an EPICS XL Flow Cytometer (Beckman Coulter).
Statistical Analyses. Statistical analyses were performed by using the unpaired Student t test.
Supplementary Material
Acknowledgments
We thank S. Yonehara, M. Ohno, H. Sakano, and H. Mitsuya for helpful discussions; A. Koito (Kumamoto University, Kumamoto, Japan) for providing the H/M/T-CD4 vector; S. Yamada (Akita University, Akita, Japan) for help in generating transgenic mice; H. Miyoshi (The Institute of Physical and Chemical Research, Tsukuba Institute, Tsukuba, Japan) for the gift of pCSII-EF-MCS vector; J. Fujisawa (Kansai Medical University, Moriguchi, Japan) for the gift of pWT-Luc; T. Hori (Kyoto University) for providing the Kit 225 cell line; and Elise Lamar for proofreading the manuscript. This study was supported by a Grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ATL, adult T cell leukemia; HBZ, HTLV-I basic leucine zipper factor; HTLV-I, human T cell leukemia virus type I; siRNA, short interfering RNA; SM HBZ, HBZ gene with silent mutations.
Data deposition: The spliced HBZ sequence reported in this paper has been deposited in the GenBank database (accession no. DQ273132).
References
- 1.Gallo, R. C. (2002) Immunol. Rev. 185, 236–265. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida, M. (2001) Annu. Rev. Immunol. 19, 475–496. [DOI] [PubMed] [Google Scholar]
- 3.Franchini, G., Fukumoto, R. & Fullen, J. R. (2003) Int. J. Hematol. 78, 280–296. [DOI] [PubMed] [Google Scholar]
- 4.Poiesz, B. J., Ruscetti, F. W., Gazdar, A. F., Bunn, P. A., Minna, J. D. & Gallo, R. C. (1980) Proc. Natl. Acad. Sci. USA 77, 7415–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallo, R. C. (2005) Retrovirology 2, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takatsuki, K. (2005) Retrovirology 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Igakura, T., Stinchcombe, J. C., Goon, P. K., Taylor, G. P., Weber, J. N., Griffiths, G. M., Tanaka, Y., Osame, M. & Bangham, C. R. (2003) Science 299, 1713–1716. [DOI] [PubMed] [Google Scholar]
- 8.Jeang, K. T., Giam, C. Z., Majone, F. & Aboud, M. (2004) J. Biol. Chem. 279, 31991–31994. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki, T., Uchida-Toita, M. & Yoshida, M. (1999) Oncogene 18, 4137–4143. [DOI] [PubMed] [Google Scholar]
- 10.Etoh, K., Tamiya, S., Yamaguchi, K., Okayama, A., Tsubouchi, H., Ideta, T., Mueller, N., Takatsuki, K. & Matsuoka, M. (1997) Cancer Res. 57, 4862–4867. [PubMed] [Google Scholar]
- 11.Cavrois, M., Leclercq, I., Gout, O., Gessain, A., Wain-Hobson, S. & Wattel, E. (1998) Oncogene 17, 77–82. [DOI] [PubMed] [Google Scholar]
- 12.Matsuoka, M. (2005) Retrovirology 2, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furukawa, Y., Kubota, R., Tara, M., Izumo, S. & Osame, M. (2001) Blood 97, 987–993. [DOI] [PubMed] [Google Scholar]
- 14.Koiwa, T., Hamano-Usami, A., Ishida, T., Okayama, A., Yamaguchi, K., Kamihira, S. & Watanabe, T. (2002) J. Virol. 76, 9389–9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda, S., Maeda, M., Morikawa, S., Taniguchi, Y., Yasunaga, J., Nosaka, K., Tanaka, Y. & Matsuoka, M. (2004) Int. J. Cancer 109, 559–567. [DOI] [PubMed] [Google Scholar]
- 16.Tamiya, S., Matsuoka, M., Etoh, K., Watanabe, T., Kamihira, S., Yamaguchi, K. & Takatsuki, K. (1996) Blood 88, 3065–3073. [PubMed] [Google Scholar]
- 17.Bangham, C. R. (2003) Int. J. Hematol. 78, 297–303. [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi, Y., Nosaka, K., Yasunaga, J. I., Maeda, M., Mueller, N., Okayama, A. & Matsuoka, M. (2005) Retrovirology 2, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larocca, D., Chao, L. A., Seto, M. H. & Brunck, T. K. (1989) Biochem. Biophys. Res. Commun. 163, 1006–1013. [DOI] [PubMed] [Google Scholar]
- 20.Gaudray, G., Gachon, F., Basbous, J., Biard-Piechaczyk, M., Devaux, C. & Mesnard, J. M. (2002) J. Virol. 76, 12813–12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basbous, J., Arpin, C., Gaudray, G., Piechaczyk, M., Devaux, C. & Mesnard, J. M. (2003) J. Biol. Chem. 278, 43620–43627. [DOI] [PubMed] [Google Scholar]
- 22.Seiki, M., Hattori, S., Hirayama, Y. & Yoshida, M. (1983) Proc. Natl. Acad. Sci. USA 80, 3618–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hori, T., Uchiyama, T., Tsudo, M., Umadome, H., Ohno, H., Fukuhara, S., Kita, K. & Uchino, H. (1987) Blood 70, 1069–1072. [PubMed] [Google Scholar]
- 24.Zuker, M. (2003) Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawada, S., Gowrishankar, K., Kitamura, R., Suzuki, M., Suzuki, G., Tahara, S. & Koito, A. (1998) J. Exp. Med. 187, 1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akagi, T., Ono, H. & Shimotohno, K. (1995) Blood 86, 4243–4249. [PubMed] [Google Scholar]
- 27.Nevins, J. R. (1998) Cell Growth Differ. 9, 585–593. [PubMed] [Google Scholar]
- 28.Weinberg, R. A. (1996) Cell 85, 457–459. [DOI] [PubMed] [Google Scholar]
- 29.Nevins, J. R. (2001) Hum. Mol. Genet. 10, 699–703. [DOI] [PubMed] [Google Scholar]
- 30.Pierce, A. M., Gimenez-Conti, I. B., Schneider-Broussard, R., Martinez, L. A., Conti, C. J. & Johnson, D. G. (1998) Proc. Natl. Acad. Sci. USA 95, 8858–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwanaga, R., Ohtani, K., Hayashi, T. & Nakamura, M. (2001) Oncogene 20, 2055–2067. [DOI] [PubMed] [Google Scholar]
- 32.Kannagi, M., Harada, S., Maruyama, I., Inoko, H., Igarashi, H., Kuwashima, G., Sato, S., Morita, M., Kidokoro, M., Sugimoto, M., et al. (1991) Int. Immunol. 3, 761–767. [DOI] [PubMed] [Google Scholar]
- 33.Inoue, J., Yoshida, M. & Seiki, M. (1987) Proc. Natl. Acad. Sci. USA 84, 3653–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicot, C., Dundr, M., Johnson, J. M., Fullen, J. R., Alonzo, N., Fukumoto, R., Princler, G. L., Derse, D., Misteli, T. & Franchini, G. (2004) Nat. Med. 10, 197–201. [DOI] [PubMed] [Google Scholar]
- 35.He, L. & Hannon, G. J. (2004) Nat. Rev. Genet. 5, 522–531. [DOI] [PubMed] [Google Scholar]
- 36.Pfeffer, S., Zavolan, M., Grasser, F. A., Chien, M., Russo, J. J., Ju, J., John, B., Enright, A. J., Marks, D., Sander, C. & Tuschl, T. (2004) Science 304, 734–736. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan, C. S., Grundhoff, A. T., Tevethia, S., Pipas, J. M. & Ganem, D. (2005) Nature 435, 682–686. [DOI] [PubMed] [Google Scholar]
- 38.Omoto, S., Ito, M., Tsutsumi, Y., Ichikawa, Y., Okuyama, H., Brisibe, E. A., Saksena, N. K. & Fujii, Y. R. (2004) Retrovirology 1, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennasser, Y., Le, S. Y., Benkirane, M. & Jeang, K. T. (2005) Immunity 22, 607–619. [DOI] [PubMed] [Google Scholar]
- 40.Iwakiri, D., Eizuru, Y., Tokunaga, M. & Takada, K. (2003) Cancer Res. 63, 7062–7067. [PubMed] [Google Scholar]
- 41.Yang, L., Aozasa, K., Oshimi, K. & Takada, K. (2004) Cancer Res. 64, 5332–5337. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida, M., Nosaka, K., Yasunaga, J., Nishikata, I., Morishita, K. & Matsuoka, M. (2004) Blood 103, 2753–2760. [DOI] [PubMed] [Google Scholar]
- 43.Bai, Y., Soda, Y., Izawa, K., Tanabe, T., Kang, X., Tojo, A., Hoshino, H., Miyoshi, H., Asano, S. & Tani, K. (2003) Gene Ther. 10, 1446–1457. [DOI] [PubMed] [Google Scholar]
- 44.Fujisawa, J., Toita, M. & Yoshida, M. (1989) J. Virol. 63, 3234–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujisawa, J., Toita, M., Yoshimura, T. & Yoshida, M. (1991) J. Virol. 65, 4525–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.