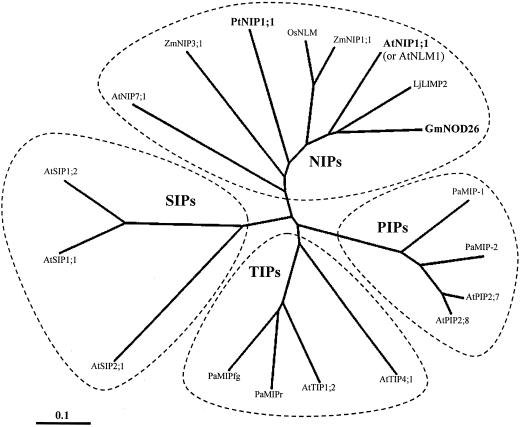
Figure 3.
Phylogenetic analysis of PtNIP1;1 with selected MIPs from the four classes: NIPs, plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPS), and small and basic intrinsic proteins (SIPs). The scale indicates nucleotide substitutions per amino acid position. Organism abbreviations are as follows: At, Arabidopsis; Gm, G. max; Lj, L. japonicus; Os, Oryza sativa; Pa, P. abies; Pt, P. taeda; Zm, Z. mays. Accession numbers are: AtNIP7;1, AAF30303; ZmNIP3;1, AF326486; PtNIP1;1, AY055751; OsNLM, BAA04257; ZmNIP1;1, AF326483; AtNIP1;1 (referred to as AtNLM1 elsewhere in text), CAA16760; LjLIMP2, AAF82791; GmNOD26, AAA02946; PaMIP-1, CAB06080; PaMIP-2, CAB07783; AtPIP2;7, CAA17774; AtPIP2;8, AAC64216; AtTIP5;1, CAB51216; AtTIP1;2, BAB01832; PaMIPr, CAA06335; PaMIPfg, CAB39758; AtSIP2;1, CAB72165; AtSIP1;1, AAF26804; and AtSIP1;2, BAB09487.