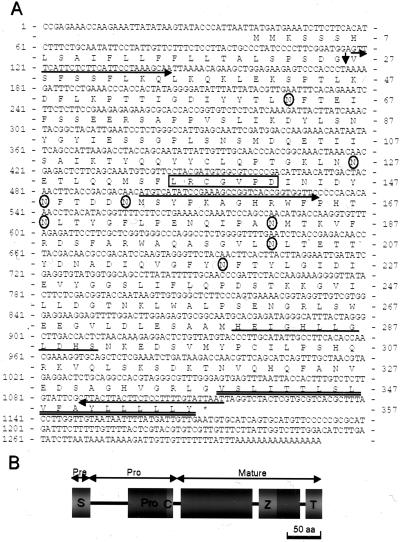
Figure 1.
Sequence analyses of the GmMMP2 cDNA and the deduced peptide. A, The deduced amino acid sequence is shown below the coding sequence, and the asterisk indicates the termination codon. The numbers at the left are for base pairs, whereas those at the right are for amino acids. A predicted signal peptide is located at amino acid position 1 through 26. The cleavage site is indicated by a vertical arrow. A forward primer denoted by the first rightward arrow right after the signal peptide below the sequence is used together with the reverse primer denoted by a leftward arrow below the sequence to generate pro-GmMMP2 sequence. Another forward primer denoted by the second rightward arrow is used together with the same reverse primer to generate the mature GmMMP2 sequence. A putative Cys switch sequence LRCGVPD is framed, whereas the zinc-binding motif is underlined. A putative C-terminal transmembrane domain is indicated by a double underline. All N residues circled by an oval are potential glycosylation sites. B, A schematic representation of the putative domains and motifs of GmMMP2. The cylinders represent those conserved domains and motifs, whereas the black lines between these domains denote the variable regions among the MMP members. S and Pre, Signal peptide; Pro, propeptide; C, Cys switch motif; Z, zinc-binding motif; T, transmembrane domain. The scale bar is equal to 50 amino acids.