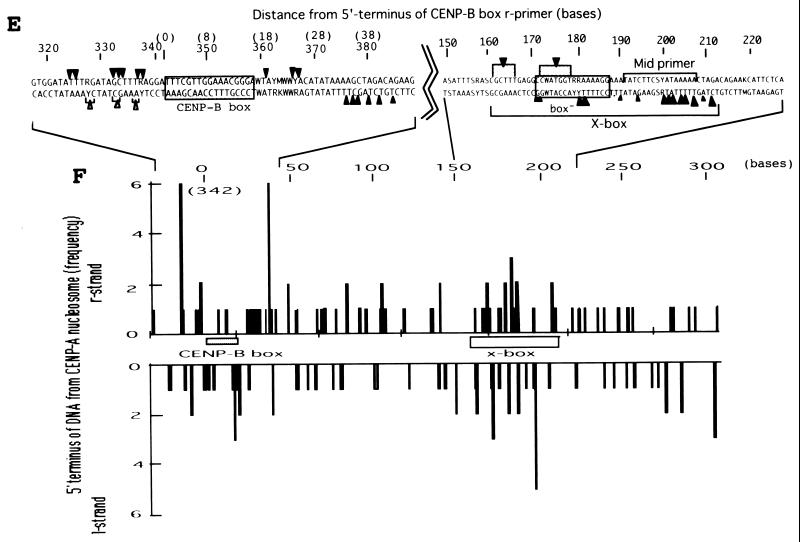
FIG. 4.
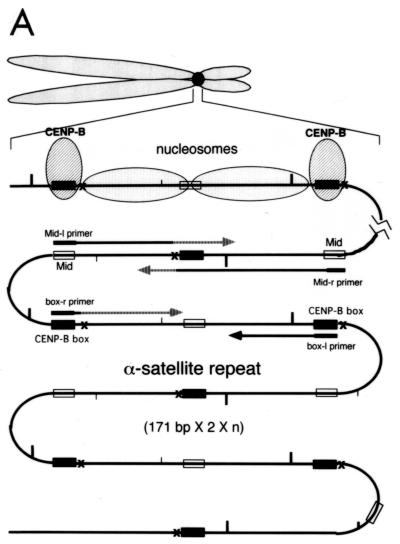
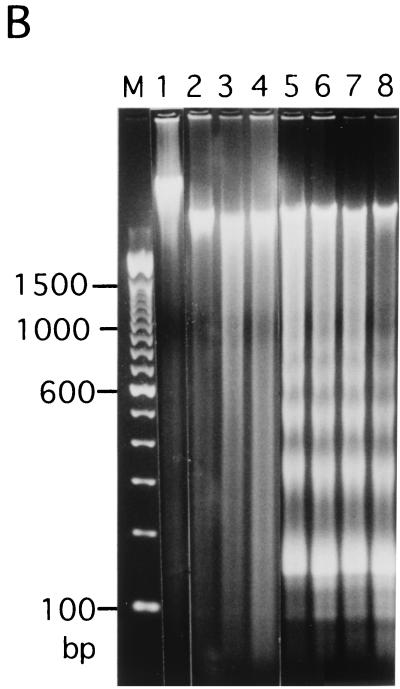
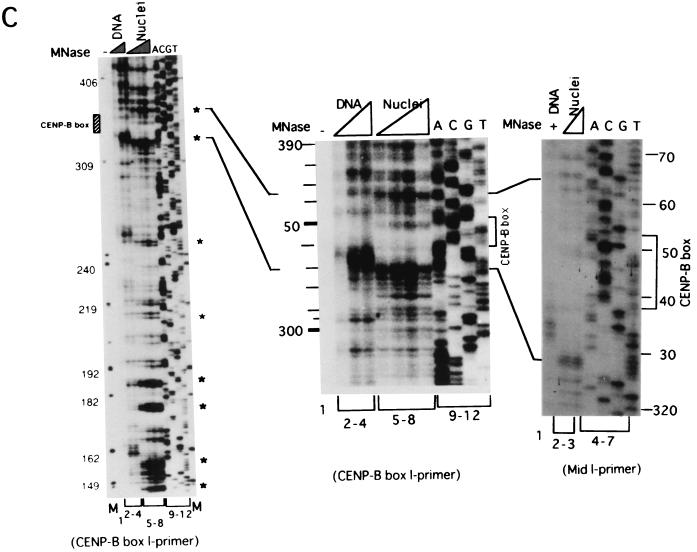
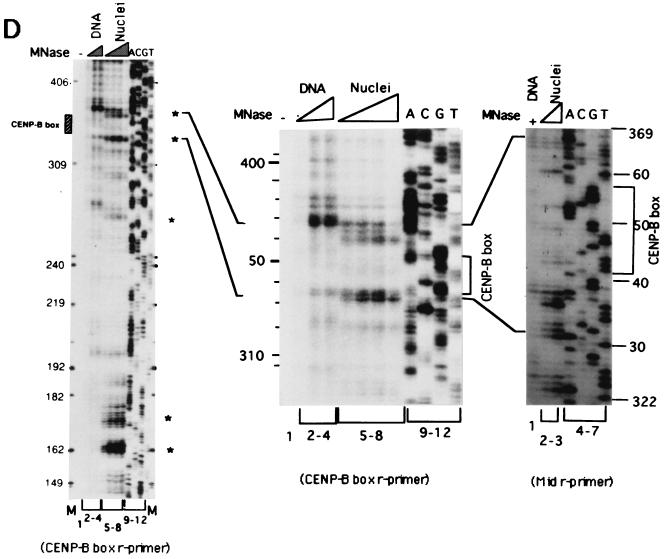
Mapping of hypersensitive sites for MNase digestion on αI-type arrays. (A) Strategy for in vivo mapping of cleavage sites by MNase digestion using CENP-B box primers or mid-primers. CENP-B boxes were located mainly in α-satellite dimer subfamilies. Dimer repeats are marked with thick vertical bars. CENP-B boxes and mid sequences are shown with solid and open boxes, respectively. The distances from the CENP-B box or the mid sequence to the hypersensitive sites for MNase digestion were measured by primer extension analyses using CENP-B boxes or mid sequences as primers. The primer for each sequence is indicated by a thick bar, and the direction of DNA synthesis is shown with arrows. Owing to the occasional deletion of 2 to 4 bases to the right of the CENP-B box (indicated by x in the figure), direct sequencing reactions were sometimes unreadable because of overlapping of the sequence ladder. (B) Agarose gel electrophoresis of HeLa DNA after MNase digestion. Purified DNA (20 μg, 0.1 ml; lanes 2 to 4) or nuclei (5 × 106, 0.1 ml; lanes 1 and 5 to 8) extracted from HeLa cells were digested with MNase (0.05 U for purified DNA and 15 U for nuclei) for 0 min (lane 1), 1 min (lane 2), 5 min (lane 3), 15 min (lane 4), 1 min (lane 5), 2 min (lane 6), 5 min (lane 7), and 20 min (lane 8). DNA from each sample was extracted with phenol and electrophoresed through a 1.7% agarose gel. Lane M shows a 100-bp DNA ladder. (C and D) PCR-primer extension analyses of the MNase digests with CENP-B box primers (left and middle panels) or mid primers (right panels). Left and middle panels, DNA (1 μg) from each of the digests shown in B was subjected to PCR-primer extension reactions using 32P-labeled CENP-B box l-primer (C) or r-primer (D), and the samples were electrophoresed for 4 h (left panels) or 6 h (middle panels). Lanes 1 to 8 correspond to the samples shown in lanes 1 to 8 of B. Lanes 9 to 12 show sequence ladders as position markers. Lane M is a 32P-labeled MspI digest of pBR322. The distance in bases from the CENP-B primer is indicated at the left. The area of the CENP-B box is marked with a shaded box, and hypersensitive bands are marked with asterisks (*). Right panels, higher-resolution analysis of the CENP-B box regions using mid l-primer (C) or r-primer (D). Lane 1 corresponds to lane 4 of B, and lanes 2 and 3 to lanes 6 and 7 of B. Lanes 4 to 7 show sequence ladders as position markers. The CENP-B box is indicated by a bracket at the right. The distance in bases from the CENP-B box is indicated at the right. (E) Mapping of MNase-hypersensitive sites around the CENP-B box and x-box regions. The DNA sequence obtained by direct sequencing of HeLa nuclei using the CENP-B box l-primer is shown (C, left, lanes 9 to 12) (61). The nucleotide position is indicated at the top as the distance in bases from the 5′ terminus of CENP-B box r-primer. Numbering around the CENP-B box region (left half of the figure) is counted from the adjacent CENP-B box. Position 342 corresponds to position 0, which is the start site for mapping around the x-box region (right half of the figure). Hypersensitive sites in the l-strand are indicated by upward-pointing arrowheads, and those in the r-strand by downward-pointing arrowheads. The upward-pointing open arrowheads with brackets at the left side of the CENP-B box indicate approximate loci (within brackets) for hypersensitive sites, because the DNA sequence of this region could not be read by direct sequencing. The CENP-B box is indicated by an open box (positions 1 to 17 or 343 to 359) and the corresponding area in the adjacent α-satellite monomer that has no affinity to CENP-B is shown as an open box (box) (positions 172 to 189). The mid-primer and x-box areas are indicated by brackets. R is A or G; Y is T or C; W is A or T; and S is G or C. (F) Mapping of 5′ termini of the α-satellite DNA fragments recovered by CHIP using anti-CENP-A antibody. 5′ termini of each strand from 94 clones of α-satellite DNA fragments (Fig. 3) were mapped on the α-satellite dimer repeat. The length of each vertical bar represents the frequency of the 5′ terminus mapping to each position on the sequence. The distance from the 5′ terminus of the CENP-B box r-strand is shown in bases at the top. Base 0 corresponds to base 342 from the adjacent CENP-B box.