FIG. 1.
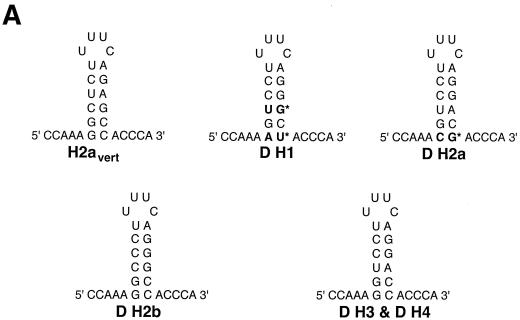
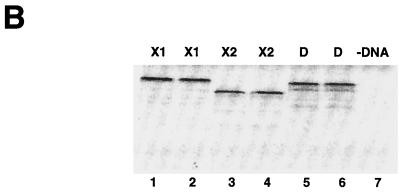
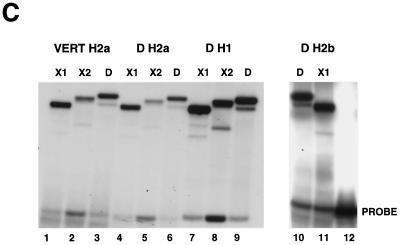
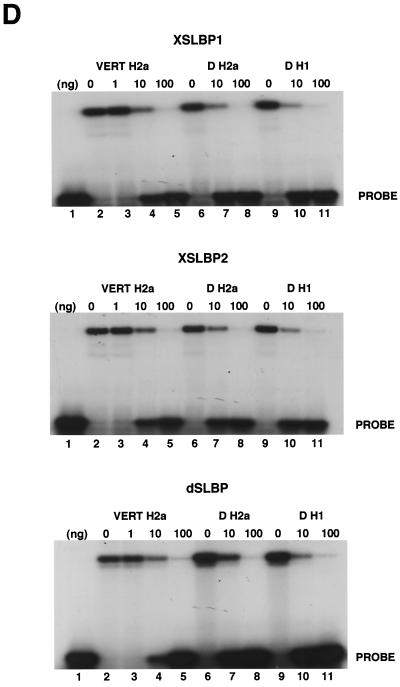
dSLBP binds to all five histone mRNAs. (A) The sequence of the 3′ end of the most common stem-loop sequence in vertebrate histone mRNAs (H2avert) and the sequence of the 3′ end of the five Drosophila histone mRNAs is shown. ∗, nucleotide pairs that are different than the canonical sequence and that have not yet been observed in any other stem-loop sequence. (B) DNAs encoding dSLBP (lanes 1 and 2), xSLBP1 (lanes 3 and 4), and xSLBP2 (lanes 5 and 6) were incubated in a reticulocyte lysate-coupled transcription-translation system in duplicate in the presence of [35S]methionine. The products were analyzed on an SDS-10% polyacrylamide gel and detected by autoradiography. Lane 7, lysate incubated without added DNA. Parallel reaction mixtures incubated in the absence of [35S]methionine were used for the mobility shift assays (C and D). (C) The indicated stem-loop sequences (lanes 1 to 3, H2avert; lanes 4 to 6, Drosophila H2a; lanes 7 to 9, Drosophila H1; lanes 10 and 11, Drosophila H2b) labeled with 32PO4 were tested for their ability to bind to Xenopus SLBP1 (X1 [lanes 1, 4, 7, and 11]), Xenopus SLBP2 (X2 [lanes 2, 5, and 8]), or Drosophila SLBP (D [lanes 3, 6, and 9]). Lane 12, probe incubated in buffer. (D) xSLBP1, xSLBP2, and dSLBP (indicated above each panel) were incubated with the H2avert probe (lanes 2 to 5), dH2a probe (lanes 6 to 8), or dH1 probe (lanes 9 to 11), and the complexes were detected by mobility shift assay. Increasing amounts of unlabeled H2avert RNA (indicated above each lane) were mixed with the probe prior to addition of the SLBPs. Lane 1, probe incubated in buffer. The complexes were analyzed by electrophoresis on an 8% polyacrylamide gel.