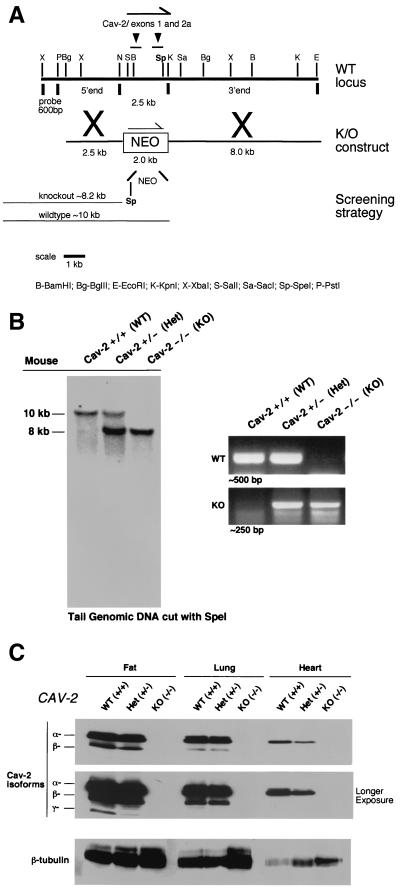
FIG. 1.
Targeted disruption of the caveolin-2 gene produces a null mutation. (A) The caveolin-2 locus (containing the first two exons) and the targeting construct (containing the neomycin resistance cassette [NEO] with flanking segments homologous to the locus) are shown in schematic format in this ≈13-kb map. The transcriptional orientations of the neomycin resistance cassette and the caveolin-2 locus are delineated by arrows. Note that homologous recombination would eliminate a 2.5-kb genomic segment containing caveolin-2 exons 1 and 2 and introduce a new restriction site (SpeI), which was used to screen for positive ES cell clones. The 600-bp XbaI-PstI probe used for Southern blot analysis is located 5′ of the targeting vector, as shown. (B) Southern blot analysis of SpeI-digested tail DNA from offspring of caveolin-2 heterozygote (Het) intercrosses. The ≈8-kb band signifiesthe appropriate targeted disruption of the caveolin-2 locus. The absence of a wild-type (WT) ≈10.0-kb band signifies the generation of the Cav-2-knockout (KO) animal. An alternative PCR-based strategy used to determine the genotype of animals is also shown. The absence of a ≈500-bp wild-type band signifies the generation of a Cav-2-knockout animal. (C) Lysates from three tissue types with various levels of caveolin-2 expression (fat, lung, and heart) were prepared for mice of all three genotypes. Protein (30 μg) was loaded in each lane, subjected to SDS-PAGE, and immunoblotted with anti-caveolin-2 MAb (clone 26). A longer exposure of the same blot is also shown. Equal protein loading was assessed by using the anti-β-tubulin MAb (clone 2.1).