Abstract
The cytokine gamma interferon (IFN-γ) and the calcitropic steroid hormone 1,25-dihydroxyvitamin D (1,25D) are activators of macrophage immune function. In sarcoidosis, tuberculosis, and several granulomatoses, IFN-γ induces 1,25D synthesis by macrophages and inhibits 1,25D induction of 24-hydroxylase, a key enzyme in 1,25D inactivation, causing high levels of 1,25D in serum and hypercalcemia. This study delineates IFN-γ-1,25D cross talk in human monocytes-macrophages. Nuclear accumulation of Stat1 and vitamin D receptor (VDR) by IFN-γ and 1,25D promotes protein-protein interactions between Stat1 and the DNA binding domain of the VDR. This prevents VDR-retinoid X receptor (RXR) binding to the vitamin D-responsive element, thus diverting the VDR from its normal genomic target on the 24-hydroxylase promoter and antagonizing 1,25D-VDR transactivation of this gene. In contrast, 1,25D enhances IFN-γ action. Stat1-VDR interactions, by preventing Stat1 deactivation by tyrosine dephosphorylation, cooperate with IFN-γ/Stat1-induced transcription. This novel 1,25D-IFN-γ cross talk explains the pathogenesis of abnormal 1,25D homeostasis in granulomatous processes and provides new insights into 1,25D immunomodulatory properties.
1,25-Dihydroxyvitamin D (1,25D), the hormonal form of vitamin D, is a potent regulator of calcium homeostasis (8). In healthy individuals, to maintain calcium concentrations within the physiological range, 1,25D tightly controls its own levels in serum (26) by dual mechanisms: suppressing its own synthesis by renal and extrarenal 1α-hydroxylase and inducing 24-hydroxylase, the key enzyme in 1,25D metabolic inactivation (27).
In contrast, in sarcoidosis, tuberculosis, several granulomatoses, and rheumatoid arthritis, abnormal 1,25D homeostasis is the cause of hypercalcemia (1, 18, 19, 34). In these patients, high levels of 1,25D in serum result from excessive 1,25D synthesis by the disease-activated macrophage (2, 3, 28) and loss of the capacity of 1,25D to regulate its own synthesis and degradation (16, 42). The demonstration of a direct correlation between pleural levels of gamma interferon (IFN-γ) and 1,25D (2) suggested the involvement of the cytokine in the abnormalities in 1,25D homeostasis. In fact, exposure of normal monocytes, pulmonary alveolar macrophages, or the monocytic cell line THP-1 to IFN-γ markedly enhances macrophage 1,25D production and antagonizes 1,25D regulation of 1α- and 24-hydroxylases (16). Clearly, in vivo and in vitro, IFN-γ impairs 1,25D control of its own synthesis and catabolism.
These studies addressed the mechanisms mediating the antagonistic effects of IFN-γ on 1,25D regulation of its own homeostasis. Since the mechanism for 1,25D-suppression of 1α-hydroxylase gene transcription is poorly understood (37), we focused on the effects of IFN-γ on 1,25D enhancement of its own catabolism. 1,25D induction of 24-hydroxylase expression is a typical 1,25D genomic action, mediated by ligand-activated vitamin D receptor (VDR) as a transcription factor (6, 12). 1,25D binding to the VDR induces a conformational change in the VDR molecule that activates the VDR to translocate to the nucleus and heterodimerize with the retinoid X receptor (RXR). The VDR-RXR complex then binds to specific DNA sequences, known as vitamin D-responsive elements (VDREs), in the 24-hydroxylase promoter (11) and recruits nuclear receptor coactivator molecules to induce transcription (8). Studies of the human monocytic cell line THP-1 demonstrated that IFN-γ may directly impair 1,25D induction of 24-hydroxylase gene transcription (16). IFN-γ inhibited 1,25D induction of 24-hydroxylase mRNA, an effect that did not result from defective binding of 1,25D to VDR or reduced stability of the 24-hydroxylase mRNA, suggesting the existence of interactions between the very distinct signaling pathways of IFN-γ and 1,25D.
In the case of IFN-γ, most responses to the cytokine involve the activation of the latent cytosolic protein Stat1 (15, 45). IFN-γ binding to its cell membrane receptor induces rapid assembly of a complete IFN-γ receptor complex with Jak1 and Jak2 enzymes, which phosphorylate one another and then phosphorylate the receptor. Receptor phosphorylation results in the formation of Stat1 docking sites. Upon phosphorylation at tyrosine 701, Stat1 homodimerizes, translocates to the nucleus, and binds DNA at specific IFN-γ activation sequences (GAS), where it either activates or represses transcription (23). Maximal transcriptional activity by active Stat1 homodimers also requires Stat1 phosphorylation at serine 727 (17, 52) and recruitment to the transcription initiation complex of the CBP-p300 family of coactivators (54).
The present study delineates the mechanism and functional relevance of the interactions between the distinct signaling pathways for the steroid hormone 1,25D and the cytokine IFN-γ. IFN-γ antagonizes 1,25D-VDR transcriptional activation of 24-hydroxylase. Direct protein-protein interactions between activated Stat1 and the DNA binding domain (DBD) of the VDR impair VDR-RXR binding to human 24-hydroxylase VDRE. In contrast, 1,25D enhances IFN-γ action. Stat1-VDR interactions prolong Stat1 activation, thus enhancing IFN-γ-Stat1 transcriptional regulation of IFN-γ-responsive genes.
MATERIALS AND METHODS
Plasmids and antibodies.
8×GAS-Luc was described previously (9, 22). Human 24-hydroxylase (−1262, +99)-chloramphenicol acetyltransferase (CAT) and monoclonal mouse anti-VDR VG1 were from H. F. DeLuca (University of Wisconsin--Madison). pSG5VDR, GST-VDR (4-427), 4× rat osteocalcin VDRE-Luc, and anti-VDR antibody 9A7 were obtained from Paul MacDonald (Case Western Reserve University, Cleveland, Ohio). GST-VDR (4-133) and GST-VDR (89-427) were provided by Mark Haussler (University of Arizona—Tucson). pET-Stat1 was obtained from Focco Van Decker (Cleveland Clinic). TGL-IP10 was obtained from Richard Ransohoff (Cleveland Clinic). LMX1B expression plasmid was obtained from Michael Rauchman (Washington University, St. Louis, Mo.). Anti-N terminus Stat1 and anti-pY701 Stat1 were purchased from Signal Transduction and New England Biolabs, respectively.
Cell culture.
THP-1 cells were grown in suspension in RPMI 1640 containing 10% fetal bovine serum. Differentiated THP-1 cells were induced to acquire a macrophage phenotype by exposure to 160 nM phorbol 12-myristate 13-acetate (Sigma, St. Louis, Mo.) for 24 h. 2fTGH, U3A, and reconstituted U3A cells (from George Stark, Cleveland Clinic) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. For all cell types, treatment with 1,25D (a gift from Milan Uskokovick, Hoffman-La Roche, Nutley, N.J.) and IFN-γ (Endogen) was conducted in serum-free medium containing 1% fatty-acid-free albumin. Incubations were done at 37°C in humidified 95% air-5% CO2.
Transfection and reporter assays.
Cells were transiently transfected by using Superfect reagent (Qiagen) according to the manufacturer's specifications. Renilla luciferase or β-galactosidase (Promega) was cotransfected as an internal standard. Firefly and Renilla luciferase, CAT, and β-galactosidase activities were all measured using Promega's kits.
Immunoprecipitation and immunoblot analysis.
THP-1, 2fTGH, or U3A cells were either untreated or stimulated with IFN-γ (1,000 IU/ml), 1,25D (50 nM), or both for 4 h. Cell extracts were prepared as described previously (30). Whole-cell extracts (600 μg or 1 mg) were immunoprecipitated with anti-VDR 9A7 or nonspecific immunoglobulin G (IgG). The immune complexes were collected on Staph-A beads (Sigma) coated with goat anti-rat antibody (Pierce) and washed four times in lysis buffer (200 mM Tris-HCl [pH 7.4], 2 mM EDTA, 0.5% Nonidet P-40, 0.3 mM sodium orthovanadate, 50 mM sodium fluoride, 0.5 mM dithiothreitol, and a cocktail of protease inhibitors [Boehringer Mannheim]). Proteins were resolved on sodium dodecyl sulfate (SDS)-7.5% polyacrylamide gels, transferred to nitrocellulose membranes, and probed with the anti-Stat1 antibody followed by anti-mouse IgG coupled to horseradish peroxidase (Bio-Rad). The transfers were analyzed by using the Supersignal chemiluminescent reagent (Pierce).
EMSAs.
Nuclear extracts or recombinant proteins were prepared, and electrophoretic mobility shift assays (EMSAs) were performed as described by Chen and DeLuca (11) or by Kotanides and Reich (29) for VDRE and GAS, respectively. The probes used were double-stranded oligonucleotides containing the 24-hydroxylase promoter-proximal VDRE sequence, 5′-ATGGAGTCAGCGAGGTGAGCGAGGGCGTCC-3′ (wild type) and 5′-ATGGAGAGTGCGAGGAGTGCGAGGAAATCC-3′ (mutant), or the high-affinity Stat binding site, SIE (m67), described by Vignais et al. (50).
GST pull-down.
The constructs for bacterial expression of glutathione S-transferase (GST) fusion proteins were transformed in Escherichia coli DH5α (Gibco-BRL). The proteins were induced and purified as described previously (13) and bound to glutathione-agarose beads (Sigma). In vitro-transcribed and -translated Stat1 and luciferase were synthesized and labeled with [35S]methionine (Redivue; Amersham Pharmacia) utilizing T7 TNT Reticulocyte Lysate Master Mix (Promega) according to the manufacturer's instructions and incubated with the GST beads. The beads were washed four times with binding buffer (20 mM Tris-HCl [pH 7.6], 50 mM NaCl, 0.2% Nonidet P-40, 1 mM dithiothreitol, and a cocktail of protease inhibitors [Boehringer Mannheim]) and two times with 50 mM Tris-HCl (pH 8.0) and eluted in 10 mM reduced glutathione-50 mM Tris-HCl, pH 8.0. The eluted fraction was resolved by SDS-polyacrylamide gel electrophoresis and analyzed using a PhosphorImager (Molecular Dynamics).
Immunofluorescence.
2fTGH or U3A cells, seeded on coverslips, were transfected with VDR and either untreated or exposed for 30 min to 1,25D (50 nM) or IFN-γ (1,000 IU/ml). The cells were fixed in methanol, blocked for 1 h in 10% goat serum-1% bovine serum albumin (BSA)-0.02% NaN3 in phosphate-buffered saline, washed once with Tris-buffered saline (TBS), and incubated overnight with anti-VDR VG1 (1:50 dilution in 1% BSA in TBS). The coverslips were washed three times in TBS, incubated with anti-mouse fluorescein isothiocyanate-conjugated antibody for 1 h in the dark followed by three washes with TBS, and mounted on slides. Images were obtained from a fluorescent confocal microscope (MRC 1024; Bio-Rad).
RNase protection assays.
Total RNA was prepared using the Trizol (Tel-Test) method according to the manufacturer's instructions. 32P-labeled riboprobes (IFN-γ-inducible protein 10 [IP-10] from Pharmingen; glyceraldehyde-3-phosphate dehydrogenase [GAPDH] from Ambion) were generated by in vitro transcription with T7 RNA polymerase (Promega). mRNAs for IP-10 and GAPDH were measured according to the manufacturer's instructions for RiboQuant (Pharmingen). Protected fragments were separated by electrophoresis on a 4.5% polyacrylamide-urea gel.
RESULTS
IFN-γ antagonizes 1,25D transcriptional activation of 24-hydroxylase.
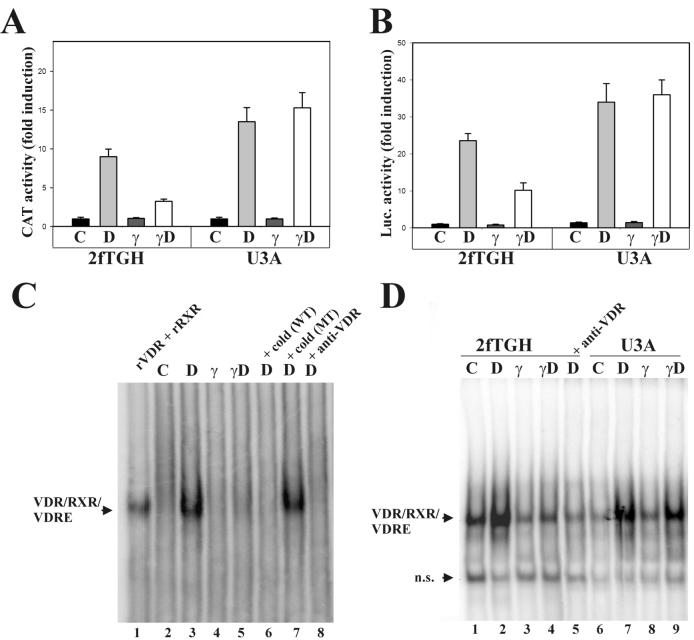
To test whether IFN-γ antagonism on 1,25D induction of 24-hydroxylase was at the transcriptional level, the human fibrosarcoma cell line 2fTGH or the Stat1-null, 2fTGH derived-U3A cells were transfected with a CAT reporter plasmid driven by the human 24-hydroxylase promoter (−1262 to +99). A β-galactosidase expression plasmid was cotransfected to normalize for transfection efficiency. In 2fTGH cells, 1,25D treatment (50 nM 1,25D) strongly induced transcription by this promoter (Fig. 1A) (11). IFN-γ (1,000 U/ml) had no effect on basal transcriptional activity; however, simultaneous treatment with 1,25D and IFN-γ reduced the transcriptional response to 1,25D by threefold. In Stat1-null U3A cells, there was no difference in transcriptional activity between cells treated with 1,25D alone and those exposed to 1,25D plus IFN-γ. These findings indicate that IFN-γ impairs 1,25D transcriptional activation of 24-hydroxylase through a Stat1-mediated mechanism.
FIG. 1.
IFN-γ antagonizes 1,25D transcriptional activity. (A) The human 24-hydroxylase promoter (−1262, +99) linked to CAT and a β-galactosidase expression plasmid were transiently transfected into wild-type 2fTGH or Stat1-null (U3A) cells. CAT activity was measured in cell lysates from untreated cells (C) or cells treated with 50 nM 1,25D (D), 1,000 U of IFN-γ/ml (γ), or both (γD) for 16 h. Results represent the means ± standard errors of the means of duplicate measurements from four independent experiments. (B) (VDRE)4-Luc and β-galactosidase expression plasmid were transiently transfected into 2fTGH or U3A cells. Luciferase activity was measured in cell lysates from cells treated as described for panel A. Results represent the means ± standard errors of the means of triplicate measurements from two independent experiments. (C) EMSA using the proximal VDRE of the human 24-hydroxylase promoter as a probe and nuclear extracts from THP-1 cells, treated for 4 h as described for panel A. Recombinant VDR and RXR (lane 1), incubation with a 300 M excess of cold wild-type (lane 6) or mutant (lane 7) VDRE, and incubation with anti-VDR 9A7 antibody (lane 8) served as controls. (D) EMSA using nuclear extracts from 2fTGH or U3A cells treated as described for panel C. n.s., nonspecific band.
VDRE is sufficient for IFN-γ antagonism.
A similar set of experiments was conducted with 2fTGH and U3A cells transiently transfected with a firefly luciferase reporter construct driven by an artificial promoter containing four copies of rat osteocalcin VDRE. As shown in Fig. 1B, this artificial promoter recapitulated the transcriptional regulation of the full-length human 24-hydroxylase promoter in the induction by 1,25D in both 2fTGH and U3A cells. Also, the antagonism on gene transcription by simultaneous treatment with IFN-γ and 1,25D persisted in 2fTGH but not U3A cells. This experiment suggests that the presence of positive consensus DR3 VDREs in a promoter is sufficient for IFN-γ antagonism on 1,25D-VDR transcriptional activation.
IFN-γ reduces VDR/RXR binding to VDRE.
IFN-γ treatment inhibited 1,25D induction of 24-hydroxylase gene transcription but did not change the basal activity of the promoter (Fig. 1A), 1,25D binding to the VDR (16), or nuclear VDR levels (data not shown). Since IFN-γ antagonism persisted in a VDRE-driven artificial promoter (Fig. 1B), the effects of the cytokine on VDR-RXR binding to VDRE were next tested. EMSAs were conducted using the proximal VDRE of the human 24-hydroxylase promoter as a probe and nuclear extracts from THP-1 cells (Fig. 1C). 1,25D treatment induced a retarded band (compare lanes 2 and 3), which corresponds to endogenous VDR-RXR bound to VDRE as judged by (i) comigration with recombinant VDR-RXR bound to VDRE (lane 1), (ii) competition by an excess of cold VDRE but not mutant VDRE (lanes 6 and 7), and (iii) inhibition of binding by incubation with anti-VDR 9A7 antibody (lane 8). VDR/RXR binding to VDRE was almost completely abolished by simultaneous treatment with 1,25D and IFN-γ (compare lanes 3 and 5). When nuclear extracts from 2fTGH cells were utilized, a similar pattern was found (Fig. 1D, lanes 1 to 4). In contrast, in Stat1-null U3A cells, there was no difference in endogenous VDR-RXR binding to VDRE between nuclear extracts from cells treated with 1,25D and extracts from cells treated with 1,25D plus IFN-γ (Fig. 1D, compare lanes 7 and 9). Taken together, these findings demonstrate that IFN-γ impairs VDR-RXR binding to VDRE through a Stat1-mediated mechanism.
VDR interacts with Stat1.
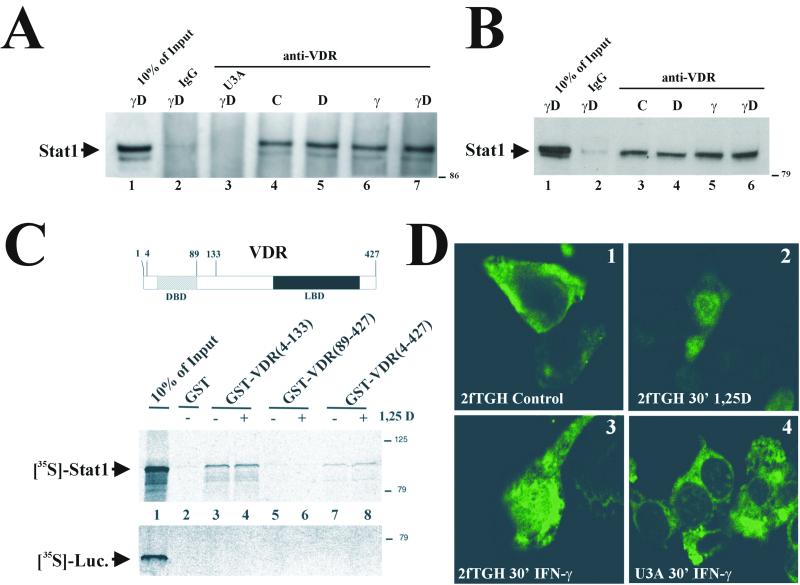
Immunoprecipitation studies examined VDR-Stat1 interactions. VDR-overexpressing 2fTGH and U3A cells were either untreated or treated with 1,25D, IFN-γ, or both for 4 h. Stat1 coprecipitated with the VDR when 2fTGH extracts were immunoprecipitated with anti-VDR but not with nonspecific IgG or when extracts from U3A cells were used (Fig. 2A). These results demonstrate that VDR and Stat1 interact in vivo. Stat1 coprecipitated with VDR in every experimental condition tested (lanes 4 to 7), indicating that VDR and Stat1 interact independently of 1,25D or IFN-γ activation.
FIG. 2.
VDR-Stat1 interaction. (A and B) Whole-cell extracts (600 μg of total protein) from VDR-transfected 2fTGH and U3A cells (A) or untransfected THP-1 (1 mg of total protein) (B) cells that were either untreated or treated with 50 nM 1,25D, 1,000 U of IFN-γ/ml, or both for 4 h were immunoprecipitated with anti-VDR 9A7 or nonspecific IgG and analyzed by Western blotting with anti-Stat1. (C) (Top) Schematic representation of the VDR. (Bottom) GST pull-down assay utilizing different GST-VDR fusion proteins purified from E. coli and bound to glutathione-agarose beads. Added was 1 μM 1,25D (+ lanes) or ethanol vehicle (− lanes). 35S-labeled Stat1 and luciferase were synthesized in vitro and incubated with the beads. After washing and elution, samples were subjected to SDS-10% polyacrylamide gel electrophoresis and autoradiographed. Numbers at right show molecular mass in kilodaltons. (D) Indirect VDR immunofluorescence in 2fTGH or U3A cells transiently transfected with VDR, either untreated or exposed to 50 nM 1,25D or 1,000 U of IFN-γ/ml for 30 min at 37°C, using a mouse monoclonal antibody against the VDR and detected with fluorescein isothiocyanate-conjugated anti-mouse antibody.
Similar results were obtained when THP-1 cells, expressing endogenous levels of VDR, were used (Fig. 2B). Clearly, IFN-γ-mediated Stat1 phosphorylation at tyrosine 701 was not required for Stat1 to interact with the VDR. Consequently, in vitro-transcribed and -translated Stat1 was tested in GST pull-down assays using different GST-VDR fusion proteins to map Stat1-VDR interactions in vitro. GST alone did not interact with Stat1 (Fig. 2C, lane 2). GST fused with full VDR (amino acids 4 to 427) interacted with Stat1 in a 1,25D-independent manner (Fig. 2C, lanes 7 and 8). A C-terminal fusion VDR, comprising amino acids 89 to 427, which contains the ligand binding domain of the VDR, did not interact with Stat1 (Fig. 2C, lanes 5 and 6). In contrast, an N-terminal fusion VDR, comprising amino acids 4 to 133, which contains the DBD, bound Stat1 with an affinity stronger than that of full-length VDR (lanes 3 and 4). In vitro-transcribed and -translated luciferase did not interact with any of the GST-VDR fusion proteins. These observations demonstrate direct interactions between Stat1 and the VDR, since no additional cellular factors were needed. The interaction maps to the DBD of the VDR, and no posttranslational modifications of Stat1, i.e., arginine 31 methylation (36) or serine 727 (52) or tyrosine 701 (43) phosphorylation, are required. The demonstration that the ligand binding domain of the VDR is not required is consistent with Stat1-VDR interaction being 1,25D independent.
IFN-γ treatment drives unliganded VDR into the nucleus.
Because Stat1-VDR interaction was ligand independent, we analyzed whether IFN-γ treatment could affect VDR subcellular localization. 2fTGH or U3A cells were transiently transfected with VDR and treated with 1,25D or IFN-γ for 30 min. The subcellular localization of the VDR was examined by indirect immunofluorescence. In both 2fTGH (Fig. 2D1 and D2) and U3A (data not shown) cells, the VDR was predominantly cytosolic in untreated cells and translocated to the nucleus within 30 min after 1,25D treatment, in agreement with previous reports (7). In 2fTGH cells treated with IFN-γ alone, a subset of the VDR was present in the nucleus in the absence of 1,25D stimulation of VDR translocation (Fig. 2D3). IFN-γ treatment was unable to translocate VDR to the nucleus in U3A cells (Fig. 2D4). Thus, IFN-γ-activated Stat1 can interact with and translocate unliganded VDR into the nucleus.
Stat1 abrogates VDR-RXR binding to VDRE.
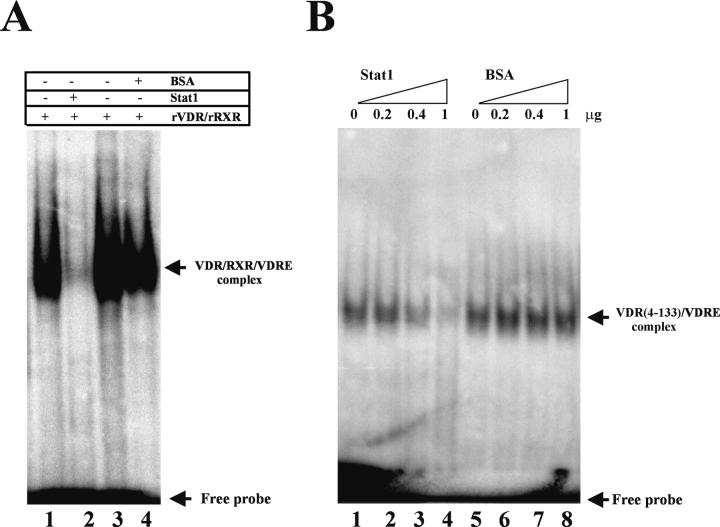
The findings that IFN-γ treatment impairs endogenous VDR-RXR binding to VDRE and that Stat1 and VDR interact in vivo and in vitro led us to determine whether Stat1 directly influenced VDR-RXR DNA binding. To this end, EMSAs were performed using the proximal VDRE of the human 24-hydroxylase promoter as a probe and recombinant proteins. Whereas VDR-RXR binding to VDRE was almost completely abolished in the presence of recombinant Stat1 (Fig. 3A, compare lanes 1 and 2), BSA, at the same concentration, had no significant effect (Fig. 3A, lanes 3 and 4).
FIG. 3.
Stat1 abrogates VDR binding to VDRE. (A) EMSA using the proximal VDRE of the human 24-hydroxylase promoter as a probe and recombinant VDR and RXR (0.1 μg each) with or without recombinant Stat1 or BSA (1 μg each). Every reaction mixture contained 1 μM 1,25D. (B) EMSA performed as described for panel A using the N-terminal (amino acids 4 to 133) portion of the VDR (0.1 μg). Binding reaction mixtures included the indicated amounts (micrograms) of Stat1 or BSA.
Since the Stat1-interacting domain of the VDR mapped to the N-terminal region containing the DBD, the possibility that Stat1 impaired VDR-DNA binding by blocking the DBD of the VDR was next tested. Figure 3B shows that the N terminus of the VDR binds VDRE, presumably as a monomer in the 3′-half site (24, 25). Stat1 inhibited the binding to DNA of this recombinant protein in a dose-dependent fashion (Fig. 3B, lanes 1 to 4). The effect was Stat1 specific, since BSA (Fig. 3B, lanes 4 to 8), tested at identical concentrations, had no effect. These results indicate that IFN-γ, by activating Stat1 and inducing its nuclear translocation, promotes Stat1 interaction with the DBD of the VDR, which blocks VDR-RXR binding to 24-hydroxylase VDRE, therefore inhibiting 1,25D induction of 24-hydroxylase gene transcription. These findings could also explain the slight but significant increase in 1,25D-induced transcription in U3A cells over that in 2fTGH cells (Fig. 1A and B). Possibly, in 2fTGH cells, small amounts of inactive Stat1 in the nucleus bind VDR-DBD, slightly decreasing 1,25D-mediated transcription, an inhibition that cannot occur in U3A cells.
1,25D has a synergistic effect on IFN-γ-mediated transcription.
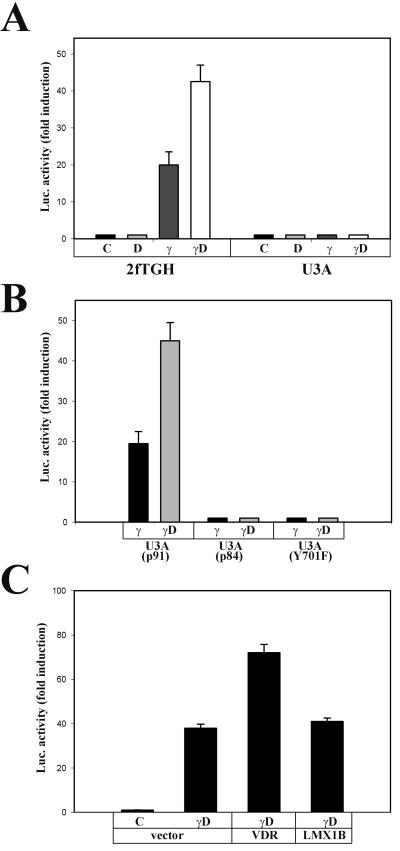
Given that Stat1-VDR interactions impaired VDR-mediated transcription, their impact on Stat1-mediated transcription was tested. 2fTGH and U3A cells were transiently transfected with a firefly luciferase reporter construct driven by a VDRE-less promoter containing eight copies of GAS. Cotransfection of a cytomegalovirus-Renilla luciferase reporter was used to assess transfection efficiency. In 2fTGH cells, IFN-γ treatment strongly induced transcription (Fig. 4A). As expected, U3A cells did not respond to IFN-γ. 1,25D, which had no effect on basal transcriptional activity by itself, enhanced IFN-γ-induced transcription. Furthermore, VDR overexpression in 2fTGH cells increased the transcription by this GAS-driven promoter, whereas the unrelated developmental transcription factor LMX1B had no effect (Fig. 4C).
FIG. 4.
1,25D synergy on IFN-γ-mediated transcription. (A) Cooperative effects of 1,25D on Stat1-induced transcription. (B) Requirements of the Stat1 molecule in 1,25D-IFN-γ synergy. (C) VDR overexpression enhances Stat1-mediated transcription. 2fTGH cells, U3A cells, and U3A cells reconstituted with different Stat1 mutants were transiently transfected with a luciferase reporter construct driven by eight copies of the consensus GAS (TTCTCGGAA) and 0.1 μg of either human VDR expression vector, human LMX1B expression vector, or vector alone when indicated. Twenty-four hours after transfection, cells were either untreated (C) or exposed to 10 nM 1,25D (D), 1,000 IU of IFN-γ/ml (γ), or both (γD) as indicated for 4 h. Luciferase activity was determined in cell lysates. Bars and error bars represent means ± standard errors of the means from triplicate measurements from two (C) or three (A and B) independent experiments.
To further characterize domains and posttranslational modifications of the Stat1 molecule necessary for 1,25D-IFN-γ synergism, experiments were conducted with U3A cells stably transfected to rescue the expression of wild-type Stat1 (p91) or different Stat1 mutants (Fig. 4B). Transcriptional synergy was rescued in U3A cells reconstituted with wild-type Stat1 (p91) but not with either a C-terminal deletion mutant lacking 7 kDa in the transactivation domain (p84) or a tyrosine phosphorylation mutant, Y701F. These results indicate that both tyrosine phosphorylation and the presence of a functional Stat1 transactivation domain are required for the synergy.
VDR prolongs Stat1 activation.
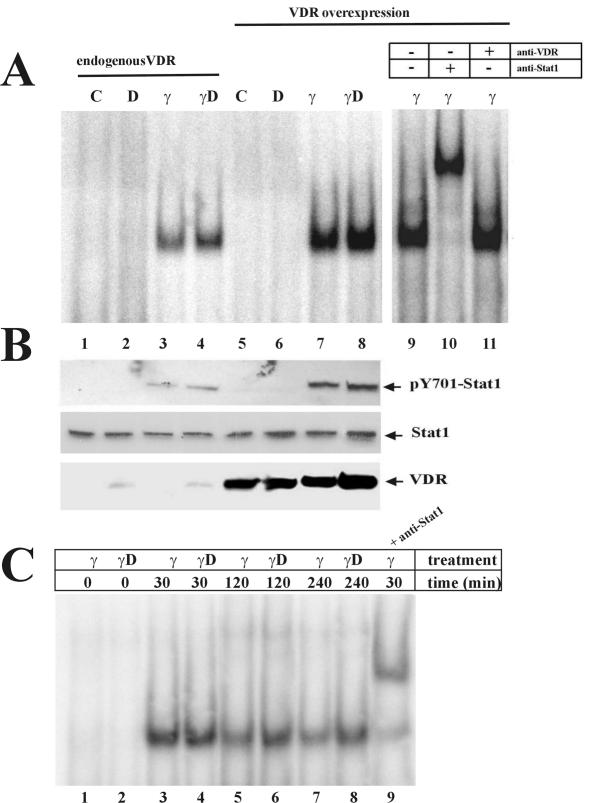
Nuclear Stat1 dephosphorylation by a yet-unknown phosphatase is one of the mechanisms by which the IFN-γ response is reduced (20, 21). The tyrosine phosphorylation of Stat1 reaches a maximum between 30 min and 1 h of IFN-γ treatment, and it starts to decrease by 2 h (38, 43). To gain some insight into potential mechanisms mediating VDR enhancement of Stat1-mediated transcription, EMSAs were conducted with a consensus GAS as probe and nuclear extracts from 2fTGH cells treated with 1,25D, IFN-γ, or both for 4 h. Simultaneous treatment with 1,25D increased Stat1 DNA binding (Fig. 5A, lanes 3 and 4). VDR overexpression further increased Stat1 binding to GAS (lanes 7 and 8). Western blot analysis showed that 1,25D treatment and/or VDR overexpression resulted in increased levels of tyrosine-phosphorylated Stat1, with no changes in total Stat1 (Fig. 5B). Time course experiments showed that simultaneous treatment with IFN-γ and 1,25D prolonged Stat1 DNA binding compared to that of IFN-γ treatment alone (Fig. 5C). These findings suggest that 1,25D-VDR could enhance IFN-γ transcriptional activity by prolonging the activated state of Stat1.
FIG. 5.
VDR prolongs Stat1 activation. (A) EMSA using a consensus GAS as a probe and nuclear extracts from untransfected 2fTGH cells or from cells transiently transfected with human VDR expression vector. Cells were either untreated or treated for 4 h with 50 nM 1,25D, 1,000 U of IFN-γ/ml, or both. The right panel shows supershift analysis utilizing anti-VDR or anti-Stat1 antibodies. (B) Western blots from cell extracts treated as described for panel A, probed with anti-pY701 Stat1, anti-Stat1, and anti-VDR. (C) EMSA analysis was done as described for panel A with cell extracts from untransfected 2fTGH cells that were treated with 1,000 U of IFN-γ/ml or 1,000 U of IFN-γ/ml plus 50 nM 1,25D for the indicated periods of time.
1,25D synergizes with IFN-γ in the induction of IP-10.
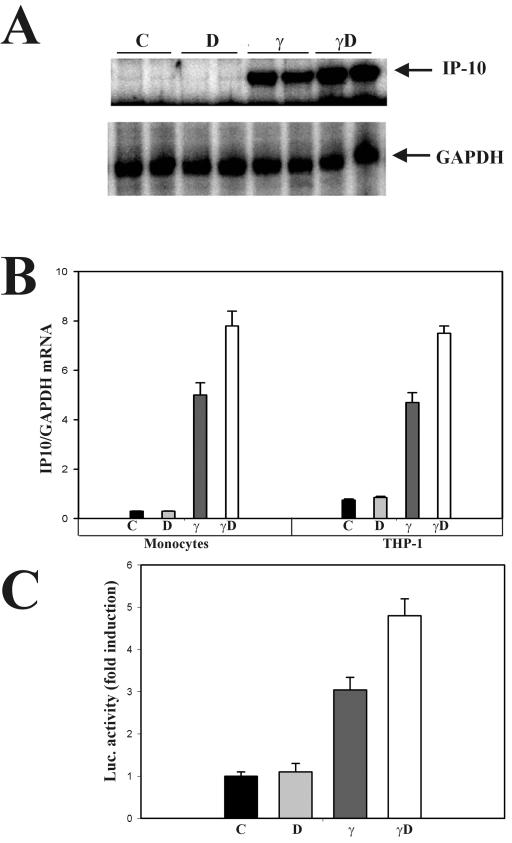
1,25D-VDR synergism on IFN-γ transcriptional activity was next tested in an endogenous gene, that for IP-10, a potent chemokine (33). RNase protection assays measured IP-10 steady-state mRNA levels in differentiated THP-1 cells (macrophage phenotype) and in peripheral blood monocytes from four healthy volunteers. In both THP-1 cells and normal monocytes, the induction of IP-10 by IFN-γ was further enhanced by simultaneous treatment with the cytokine and 1,25D (Fig. 6A, left and right panels, respectively). Densitometric quantitation of IP-10/GAPDH mRNA ratios showed that simultaneous treatment with 1,25D and IFN-γ increased by 60% the induction of IP-10 mRNA by IFN-γ alone (Fig. 6B). Τransient-transfection studies of 2fTGH cells with a luciferase reporter construct driven by the human IP-10 promoter recapitulated the regulation found in mRNA levels (Fig. 6C). This synergy in the induction of IP-10 indicates that the effects observed in a GAS-driven artificial promoter take place, indeed, in an endogenous promoter.
FIG. 6.
1,25D synergy on IFN-γ induction of the IP-10 gene. (A) Normal blood monocytes were left untreated or treated with 50 nM 1,25D, 150 U of IFN-γ/ml, or both for 18 h. IP-10 and GAPDH mRNAs were measured by RNase protection assays. (B) Relative IP-10/GAPDH mRNA levels in peripheral blood monocytes from four healthy volunteers (left graph) or differentiated THP-1 cells (right graph) from two independent experiments. Results represent the means ± standard errors of the means. (C) 2fTGH cells were transiently transfected with TGL-IP10. Twenty-four hours after transfection, cells were either untreated (C) or exposed to 10 nM 1,25D (D), 500 IU of IFN-γ/ml (γ), or both (γD) as indicated for 24 h. Luciferase activity was determined in cell lysates. Bars and error bars represent means ± standard errors of the means from triplicate measurements.
DISCUSSION
The present study, designed to characterize the mechanisms mediating IFN-γ antagonism on 1,25D control of its own homeostasis, demonstrates a novel cross talk between the distinct signaling pathways of the cytokine IFN-γ and the hormonal form of vitamin D. Direct protein-protein interactions between Stat1 and the VDR, the transcription factors for IFN-γ and 1,25D, inhibit 1,25D transcriptional activity and enhance Stat1-mediated transcription.
In vitro studies using recombinant proteins addressed the mechanisms for IFN-γ inhibition of 1,25D transcriptional activity, demonstrating that (i) Stat1-VDR interactions also occur in the absence of ligand-mediated activation of either protein, (ii) Stat1 binds the DBD of the VDR, and (iii) Stat1 binding to the DBD of the VDR is sufficient to reduce VDR binding to VDRE. The biological relevance of these findings was confirmed in vivo. Coimmunoprecipitation experiments with THP-1 and 2fTGH cells showed that endogenous nuclear Stat1 and the VDR physically interact. In both cell types, Stat1 binds the VDR, reducing VDR-RXR binding to the 24-hydroxylase VDRE. In 2fTGH cells, IFN-γ-induced reduction in VDR/RXR binding to VDRE resulted in decreased 1,25D-VDR transcriptional activation of the 24-hydroxylase gene. In Stat1-null U3A cells, neither VDR-RXR binding to VDRE nor 1,25D transactivation of 24-hydroxylase was impaired by IFN-γ treatment. Since both proximal and distal VDREs in the human 24-hydroxylase promoter are typical VDRE sequences, inhibitory effects of IFN-γ identical to those exerted on 1,25D transactivation of human 24-hydroxylase could affect other genes induced by 1,25D through classical VDREs. In fact, IFN-γ antagonized 1,25D induction of a rat osteocalcin VDRE-driven reporter (Fig. 1B). Thus, the mechanisms described by these studies may explain IFN-γ antagonism on 1,25D induction of the osteocalcin gene (39) and IFN-γ reduction of bone formation (44).
In contrast to the inhibitory effects of Stat1-VDR interactions on 1,25D action, 1,25D has cooperative effects on IFN-γ 47-Stat1 transcriptional activation. Specifically, 1,25D enhanced IFN-γ induction of the mRNA levels of the potent chemokine IP-10 in normal human monocytes and in differentiated THP-1 cells (macrophage phenotype), as well as IFN-γ transactivation of a human IP-10 promoter-driven reporter. 1,25D synergy on Stat1-induced transcription was reproduced in 2fTGH cells in a GAS-driven, VDRE-less artificial promoter. VDR-Stat1 synergy increased with VDR cotransfection.
Cross talk between Stat proteins and steroid receptors has been reported. Similar to the functional interactions between Stat1 and the VDR, Stat5 antagonizes transactivation of glucocorticoid receptor (GR)-responsive promoters through direct protein-protein interactions that also result in GR synergism on Stat5-mediated transcription of the β-casein gene (47). Further characterization of GR-Stat5 interactions revealed the DBD of the GR as dispensable for cooperation with Stat5, whereas the AF1 transactivation domain is required, thus suggesting that the GR acts as a ligand-dependent coactivator of Stat5. In contrast, the Stat5 transactivation domain is not required for GR cooperation (48).
Diverse cross-modulations occur between Stats and steroid receptors. Stat5 also antagonizes transcription by mineralocorticoid, progesterone, and estrogen receptors. However, only mineralocorticoid and progesterone receptors enhance Stat5-mediated transcription, which is inhibited by the estrogen receptor and unaffected by the androgen receptor (49). In contrast to Stat5 antagonism on transactivation by steroid receptors, interleukin-6-activated Stat3 acts as a GR transcriptional coactivator (55).
Physical Stat-steroid receptor interactions are not mandatory for cross talk between their cognate signaling pathways, since indirect mechanisms of cross-modulation also exist. In fact, there is no protein-protein interaction mediating Stat5b inhibition of peroxisome proliferator-activated receptor α transcriptional activity (56), GR enhancement of Stat1 activation of Fcγ receptor I in monocytes (4), or retinoic acid synergy on IFN-γ action (10).
The demonstration that 1,25D treatment and VDR overexpression markedly enhance Stat1 binding to GAS suggested that the VDR could operate as a coactivator of Stat1-mediated transcription. Although the possibility that active Stat1-VDR-containing complexes bind GAS in vivo cannot be completely ruled out, no complexes containing VDR and Stat1 bound to GAS were detected when supershifting EMSAs with an antibody against the VDR, even when overexpressing the VDR.
Tyrosine dephosphorylation of Stat1, by a yet-unknown nuclear phosphatase, deactivates IFN-γ signaling (20, 21). In fact, the vaccinia virus blocks IFN-γ action by dephosphorylating Stat1 through a virion-encoded phosphatase (38). Since 1,25D treatment prolonged the activated state of Stat1, we propose that Stat1-VDR interactions, by protecting Stat1 from inactivation by tyrosine dephosphorylation, enhance Stat1 binding to GAS and, consequently, Stat1-mediated induction of IFN-γ-regulated genes. Similarly, the GR prolongs the activated state of Stat5 (53), thus enhancing Stat5-mediated transcription.
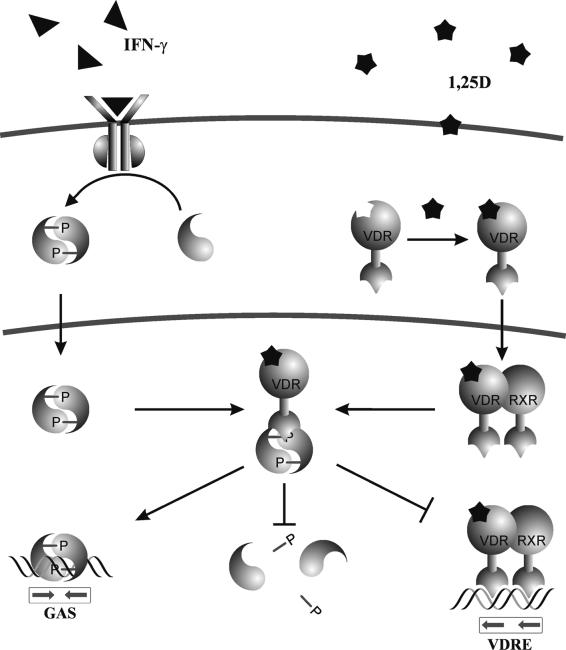
In summary, among numerous potential mechanisms, our data suggest that, in human monocytes and macrophages, functional Stat1-VDR interactions result from the cross-modulation depicted in Fig. 7. Simultaneous exposure to 1,25D and IFN-γ increases nuclear VDR and Stat1 levels, thus promoting direct protein-protein interactions between the two transcription factors that prevent VDR-RXR binding to the VDRE and deactivation of Stat1 by tyrosine dephosphorylation. The biological consequences of nuclear VDR-Stat1 complex formation include IFN-γ antagonism on 1,25D-VDR transcriptional activity and 1,25D enhancement of IFN-γ-Stat1-induced transcription.
FIG. 7.
Schematic diagram of functional Stat1-VDR interactions. Nuclear accumulation of Stat1 and the VDR by IFN-γ and 1,25D promotes physical interaction between Stat1 and the DBD of the VDR. Stat1-VDR complex formation inhibits VDR-RXR binding to VDRE and Stat1 deactivation by tyrosine dephosphorylation, thus resulting in IFN-γ antagonism on 1,25D-VDR transactivation and 1,25D cooperation on Stat1-mediated transcription.
This model explains the pathogenesis of the abnormal 1,25D homeostasis and the resultant hypercalcemia in granulomatous processes. High concentrations of circulating 1,25D result from Stat1-VDR antagonism on 1,25D transactivation of 24-hydroxylase, the key enzyme for 1,25D inactivation. Reduction of 24-hydroxylase itself causes increased levels of 1,25D in serum (46). Also, Stat1-VDR synergy on IFN-γ action may contribute to increase serum 1,25D levels. In fact, 1α-hydroxylase mRNA levels in differentiated THP-1 cells exposed to 1,25D and IFN-γ are 45% higher than the levels in cells treated with IFN-γ alone (35).
Although the influence of different promoter architectures and/or cellular patterns of protein expression advises caution before generalizing the impact of this model to every 1,25D- and IFN-γ-regulated gene, VDR-Stat1 interactions may also provide new insights into the immunomodulatory properties of 1,25D.
1,25D is a potent immunosuppressor (31). The 1,25D produced by the activated macrophage exerts paracrine effects on surrounding activated T lymphocytes, inhibiting their production of interleukin-2 (5) and IFN-γ itself (14). Thus, 1,25D mediates a negative feedback loop to decrease inflammation (32). IFN-γ-induced Stat1-VDR antagonism on 1,25D action on macrophages, which cause high levels of 1,25D, is, however, unlikely to affect TH1 lymphocytes, the main type responsible for inflammatory responses, because these cells lack the IFN-γ receptor β chain (41), mandatory for IFN-γ action.
VDR-Stat1 synergy may further enhance immunomodulation by 1,25D on monocytes/macrophages. In fact, synergistic effects of 1,25D and IFN-γ were reported previously for the induction of nonspecific esterase (51) and NADPH oxidase (its regulated subunits) (40) genes, two processes directly associated with terminal macrophage differentiation and enhanced antimicrobicidal and antiviral potential. Since both 1,25D and IFN-γ are activators of macrophage immune function, high levels of 1,25D could exert autocrine effects in activated macrophages by synergizing with IFN-γ, the most potent macrophage-activating cytokine.
Acknowledgments
This work was supported by grant AR 45283 from the National Institutes of Health (to A.S.D.).
We thank Paul MacDonald and Mark Haussler for their generous contributions to our research, Alejandro Barbieri and Julia B. Cordero for confocal microscopy imaging, Richard Ransohoff for his prompt response in providing the construct TGL-IP10, Adrian Arakaki for the design of the diagram model, and Zhengmin Huang for his assistance in transfection assays.
REFERENCES
- 1.Adams, J. S., M. A. Gacad, A. Anders, D. B. Endres, and O. P. Sharma. 1986. Biochemical indicators of disordered vitamin D and calcium homeostasis in sarcoidosis. Sarcoidosis 3:1-6. [PubMed] [Google Scholar]
- 2.Adams, J. S., R. L. Modlin, M. M. Diz, and P. F. Barnes. 1989. Potentiation of the macrophage 25-hydroxyvitamin D-1-hydroxylation reaction by human tuberculous pleural effusion fluid. J. Clin. Endocrinol. Metab. 69:457-460. [DOI] [PubMed] [Google Scholar]
- 3.Adams, J. S., F. R. Singer, M. A. Gacad, O. P. Sharma, M. J. Hayes, P. Vouros, and M. F. Holick. 1985. Isolation and structural identification of 1,25-dihydroxyvitamin D3 produced by cultured alveolar macrophages in sarcoidosis. J. Clin. Endocrinol. Metab. 60:960-966. [DOI] [PubMed] [Google Scholar]
- 4.Aittomaki, S., M. Pesu, B. Groner, O. A. Janne, J. J. Palvimo, and O. Silvennoinen. 2000. Cooperation among Stat1, glucocorticoid receptor, and PU.1 in transcriptional activation of the high-affinity Fc gamma receptor I in monocytes. J. Immunol. 164:5689-5697. [DOI] [PubMed] [Google Scholar]
- 5.Alroy, I., T. L. Towers, and L. P. Freedman. 1995. Transcriptional repression of the interleukin-2 gene by vitamin D3: direct inhibition of NFATp/AP-1 complex formation by a nuclear hormone receptor. Mol. Cell. Biol. 15:5789-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armbrecht, H. J., T. L. Hodam, M. A. Boltz, and M. L. Chen. 1993. Phorbol ester markedly increases the sensitivity of intestinal epithelial cells to 1,25-dihydroxyvitamin D3. FEBS Lett. 327:13-16. [DOI] [PubMed] [Google Scholar]
- 7.Barsony, J., J. W. Pike, H. F. DeLuca, and S. J. Marx. 1990. Immunocytology with microwave-fixed fibroblasts shows 1α,25-dihydroxyvitamin D3-dependent rapid and estrogen-dependent slow reorganization of vitamin D receptors. J. Cell Biol. 111:2385-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, A. J., A. Dusso, and E. Slatopolsky. 1999. Vitamin D. Am. J. Physiol. 277:F157-F175. [DOI] [PubMed]
- 9.Chatterjee-Kishore, M., K. L. Wright, J. P. Ting, and G. R. Stark. 2000. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 19:4111-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chelbi-Alix, M. K., and L. Pelicano. 1999. Retinoic acid and interferon signaling cross talk in normal and RA-resistant APL cells. Leukemia 13:1167-1174. [DOI] [PubMed] [Google Scholar]
- 11.Chen, K. S., and H. F. DeLuca. 1995. Cloning of the human 1α,25-dihydroxyvitamin D-3 24-hydroxylase gene promoter and identification of two vitamin D-responsive elements. Biochim. Biophys. Acta 1263:1-9. [DOI] [PubMed] [Google Scholar]
- 12.Chen, M. L., M. A. Boltz, and H. J. Armbrecht. 1993. Effects of 1,25-dihydroxyvitamin D3 and phorbol ester on 25-hydroxyvitamin D3 24-hydroxylase cytochrome P450 messenger ribonucleic acid levels in primary cultures of rat renal cells. Endocrinology 132:1782-1788. [DOI] [PubMed] [Google Scholar]
- 13.Chen, N., T. Baudino, P. N. MacDonald, M. Green, W. L. Kelley, J. W. Burnett, and R. M. Buller. 2000. Selective inhibition of nuclear steroid receptor function by a protein from a human tumorigenic poxvirus. Virology 274:17-25. [DOI] [PubMed] [Google Scholar]
- 14.Cippitelli, M., and A. Santoni. 1998. Vitamin D3: a transcriptional modulator of the interferon-gamma gene. Eur. J. Immunol. 28:3017-3030. [DOI] [PubMed] [Google Scholar]
- 15.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 16.Dusso, A. S., S. Kamimura, M. Gallieni, M. Zhong, L. Negrea, S. Shapiro, and E. Slatopolsky. 1997. γ-Interferon-induced resistance to 1,25-(OH)2 D3 in human monocytes and macrophages: a mechanism for the hypercalcemia of various granulomatoses. J. Clin. Endocrinol. Metab. 82:2222-2232. [DOI] [PubMed] [Google Scholar]
- 17.Eilers, A., D. Georgellis, B. Klose, C. Schindler, A. Ziemiecki, A. G. Harpur, A. F. Wilks, and T. Decker. 1995. Differentiation-regulated serine phosphorylation of STAT1 promotes GAF activation in macrophages. Mol. Cell. Biol. 15:3579-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gates, S., J. Shary, R. T. Turner, S. Wallach, and N. H. Bell. 1986. Abnormal calcium metabolism caused by increased circulating 1,25-dihydroxyvitamin D in a patient with rheumatoid arthritis. J. Bone Miner. Res. 1:221-226. [DOI] [PubMed] [Google Scholar]
- 19.Gkonos, P. J., R. London, and E. D. Hendler. 1984. Hypercalcemia and elevated 1,25-dihydroxyvitamin D levels in a patient with end-stage renal disease and active tuberculosis. N. Engl. J. Med. 311:1683-1685. [DOI] [PubMed] [Google Scholar]
- 20.Haspel, R. L., and J. E. Darnell, Jr. 1999. A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl. Acad. Sci. USA 96:10188-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haspel, R. L., M. Salditt-Georgieff, and J. E. Darnell, Jr. 1996. The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. EMBO J. 15:6262-6268. [PMC free article] [PubMed] [Google Scholar]
- 22.Horvai, A. E., L. Xu, E. Korzus, G. Brard, D. Kalafus, T. M. Mullen, D. W. Rose, M. G. Rosenfeld, and C. K. Glass. 1997. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc. Natl. Acad. Sci. USA 94:1074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath, C. M., and J. E. Darnell. 1997. The state of the STATs: recent developments in the study of signal transduction to the nucleus. Curr. Opin. Cell Biol. 9:233-239. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh, J. C., S. Nakajima, M. A. Galligan, P. W. Jurutka, C. A. Haussler, G. K. Whitfield, and M. R. Haussler. 1995. Receptor mediated genomic action of the 1,25(OH)2D3 hormone: expression of the human vitamin D receptor in E. coli. J. Steroid Biochem. Mol. Biol. 53:583-594. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh, J. C., G. K. Whitfield, A. K. Oza, H. T. Dang, J. N. Price, M. A. Galligan, P. W. Jurutka, P. D. Thompson, C. A. Haussler, and M. R. Haussler. 1999. Characterization of unique DNA-binding and transcriptional-activation functions in the carboxyl-terminal extension of the zinc finger region in the human vitamin D receptor. Biochemistry 38:16347-16358. [DOI] [PubMed] [Google Scholar]
- 26.Hughes, M. R., D. J. Baylink, P. G. Jones, and M. R. Haussler. 1976. Radioligand receptor assay for 25-hydroxyvitamin D2/D3 and 1α,25-dihydroxyvitamin D2/D3. J. Clin. Investig. 58:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knutson, J. C., and H. F. DeLuca. 1974. 25-Hydroxyvitamin D3-24-hydroxylase. Subcellular location and properties. Biochemistry 13:1543-1548. [DOI] [PubMed] [Google Scholar]
- 28.Koeffler, H. P., H. Reichel, J. E. Bishop, and A. W. Norman. 1985. γ-Interferon stimulates production of 1,25-dihydroxyvitamin D3 by normal human macrophages. Biochem. Biophys. Res. Commun. 127:596-603. [DOI] [PubMed] [Google Scholar]
- 29.Kotanides, H., and N. C. Reich. 1993. Requirement of tyrosine phosphorylation for rapid activation of a DNA binding factor by IL-4. Science 262:1265-1267. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, A., Y. L. Yang, V. Flati, S. Der, S. Kadereit, A. Deb, J. Haque, L. Reis, C. Weissmann, and B. R. Williams. 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 16:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemire, J. 2000. 1,25-Dihydroxyvitamin D3--a hormone with immunomodulatory properties. Z. Rheumatol. 59:24-27. [DOI] [PubMed] [Google Scholar]
- 32.Lemire, J. M., D. C. Archer, L. Beck, and H. L. Spiegelberg. 1995. Immunosuppressive actions of 1,25-dihydroxyvitamin D3: preferential inhibition of Th1 functions. J. Nutr. 125:1704S-1708S. [DOI] [PubMed] [Google Scholar]
- 33.Luster, A. D., J. C. Unkeless, and J. V. Ravetch. 1985. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 315:672-676. [DOI] [PubMed] [Google Scholar]
- 34.Meyrier, A., D. Valeyre, R. Bouillon, F. Paillard, J. P. Battesti, and R. Georges. 1985. Resorptive versus absorptive hypercalciuria in sarcoidosis: correlations with 25-hydroxy vitamin D3 and 1,25-dihydroxy vitamin D3 and parameters of disease activity. Q. J. Med. 54:269-281. [PubMed] [Google Scholar]
- 35.Monkawa, T., T. Yoshida, M. Hayashi, and T. Saruta. 2000. Identification of 25-hydroxyvitamin D3 1α-hydroxylase gene expression in macrophages. Kidney Int. 58:559-568. [DOI] [PubMed] [Google Scholar]
- 36.Mowen, K. A., J. Tang, W. Zhu, B. T. Schurter, K. Shuai, H. R. Herschman, and M. David. 2001. Arginine methylation of STAT1 modulates IFNα/β-induced transcription. Cell 104:731-741. [DOI] [PubMed] [Google Scholar]
- 37.Murayama, A., K. Takeyama, S. Kitanaka, Y. Kodera, Y. Kawaguchi, T. Hosoya, and S. Kato. 1999. Positive and negative regulations of the renal 25-hydroxyvitamin D3 1α-hydroxylase gene by parathyroid hormone, calcitonin, and 1α,25(OH)2D3 in intact animals. Endocrinology 140:2224-2231. [DOI] [PubMed] [Google Scholar]
- 38.Najarro, P., P. Traktman, and J. A. Lewis. 2001. Vaccinia virus blocks gamma interferon signal transduction: viral VH1 phosphatase reverses Stat1 activation. J. Virol. 75:3185-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanes, M. S., J. Rubin, L. Titus, G. N. Hendy, and B. D. Catherwood. 1990. Interferon-gamma inhibits 1,25-dihydroxyvitamin D3-stimulated synthesis of bone GLA protein in rat osteosarcoma cells by a pretranslational mechanism. Endocrinology 127:588-594. [DOI] [PubMed] [Google Scholar]
- 40.Obermeier, H., A. Sellmayer, U. Danesch, and M. Aepfelbacher. 1995. Cooperative effects of interferon-gamma on the induction of NADPH oxidase by retinoic acid or 1,25(OH)2-vitamin D3 in monocytic U937 cells. Biochim. Biophys. Acta 1269:25-31. [DOI] [PubMed] [Google Scholar]
- 41.Pernis, A., S. Gupta, K. J. Gollob, E. Garfein, R. L. Coffman, C. Schindler, and P. Rothman. 1995. Lack of interferon gamma receptor beta chain and the prevention of interferon gamma signaling in TH1 cells. Science 269:245-247. [DOI] [PubMed] [Google Scholar]
- 42.Reichel, H., H. P. Koeffler, R. Barbers, and A. W. Norman. 1987. Regulation of 1,25-dihydroxyvitamin D3 production by cultured alveolar macrophages from normal human donors and from patients with pulmonary sarcoidosis. J. Clin. Endocrinol. Metab. 65:1201-1209. [DOI] [PubMed] [Google Scholar]
- 43.Shuai, K., C. Schindler, V. R. Prezioso, and J. E. Darnell, Jr. 1992. Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258:1808-1812. [DOI] [PubMed] [Google Scholar]
- 44.Smith, D. D., M. Gowen, and G. R. Mundy. 1987. Effects of interferon-gamma and other cytokines on collagen synthesis in fetal rat bone cultures. Endocrinology 120:2494-2499. [DOI] [PubMed] [Google Scholar]
- 45.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 46.St.-Arnaud, R., A. Arabian, R. Travers, F. Barletta, M. Raval-Pandya, K. Chapin, J. Depovere, C. Mathieu, S. Christakos, M. B. Demay, and F. H. Glorieux. 2000. Deficient mineralization of intramembranous bone in vitamin D-24-hydroxylase-ablated mice is due to elevated 1,25-dihydroxyvitamin D and not to the absence of 24,25-dihydroxyvitamin D. Endocrinology 141:2658-2666. [DOI] [PubMed] [Google Scholar]
- 47.Stocklin, E., M. Wissler, F. Gouilleux, and B. Groner. 1996. Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383:726-728. [DOI] [PubMed] [Google Scholar]
- 48.Stoecklin, E., M. Wissler, R. Moriggl, and B. Groner. 1997. Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol. Cell. Biol. 17:6708-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoecklin, E., M. Wissler, D. Schaetzle, E. Pfitzner, and B. Groner. 1999. Interactions in the transcriptional regulation exerted by Stat5 and by members of the steroid hormone receptor family. J. Steroid Biochem. Mol. Biol. 69:195-204. [DOI] [PubMed] [Google Scholar]
- 50.Vignais, M. L., H. B. Sadowski, D. Watling, N. C. Rogers, and M. Gilman. 1996. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol. Cell. Biol. 16:1759-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinberg, J. B., M. A. Misukonis, M. M. Hobbs, and M. J. Borowitz. 1986. Cooperative effects of gamma interferon and 1-α,25-dihydroxyvitamin D3 in inducing differentiation of human promyelocytic leukemia (HL-60) cells. Exp. Hematol. 14:138-142. [PubMed] [Google Scholar]
- 52.Wen, Z., Z. Zhong, and J. E. Darnell, Jr. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241-250. [DOI] [PubMed] [Google Scholar]
- 53.Wyszomierski, S. L., J. Yeh, and J. M. Rosen. 1999. Glucocorticoid receptor/signal transducer and activator of transcription 5 (STAT5) interactions enhance STAT5 activation by prolonging STAT5 DNA binding and tyrosine phosphorylation. Mol. Endocrinol. 13:330-343. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, J. J., U. Vinkemeier, W. Gu, D. Chakravarti, C. M. Horvath, and J. E. Darnell, Jr. 1996. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc. Natl. Acad. Sci. USA 93:15092-15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Z., S. Jones, J. S. Hagood, N. L. Fuentes, and G. M. Fuller. 1997. STAT3 acts as a co-activator of glucocorticoid receptor signaling. J. Biol. Chem. 272:30607-30610. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, Y. C., and D. J. Waxman. 1999. Cross-talk between janus kinase-signal transducer and activator of transcription (JAK-STAT) and peroxisome proliferator-activated receptor-alpha (PPARα) signaling pathways. Growth hormone inhibition of pparα transcriptional activity mediated by stat5b. J. Biol. Chem. 274:2672-2681. [DOI] [PubMed] [Google Scholar]