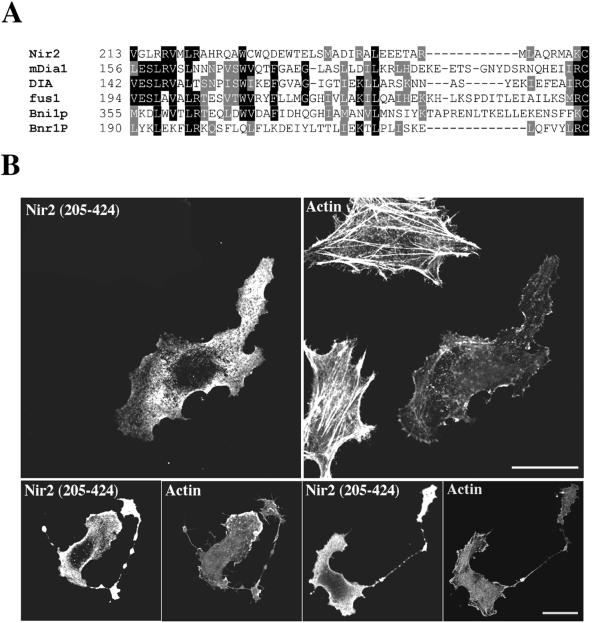
FIG. 2.
The N-terminal region of Nir2 contains a Rid. (A) Sequence alignment of Nir2 and the N-terminal region of several FH proteins. The different protein sequences used in this alignment are for Nir2 (Homo sapiens, GenBank accession no. AF334584), mDia1 (Mus musculus; GenBank accession no. U96963), DIA (D. melanogaster, GenBank accession no. U11288), fus1 (Schizosaccharomyces pombe, GenBank accession no. Z71547), Bni1p (Saccharomyces cerevisiae, GenBank accession no. Z71547), and Bnr1P (S. cerevisiae, GenBank accession no. Z47047). Identical residues among at least three of the aligned sequences are boxed in black, and residues representing conserved substitutions are boxed in gray. Numbers indicate the position of the first residue relative to the amino acid sequence of the indicated proteins. Sequences were aligned using the ClustalW algorithm. (B) Effect of Nir2 variant on the organization of the actin cytoskeleton. A short variant of Nir2 (aa 205 to 424) was expressed in HeLa cells as an HA-tagged fusion protein, and its effect on F-actin and cell morphology was analyzed by fluorescence microscopy. Shown are representative phenotypes of the transfected cells stained with anti-HA antibodies and the filamentous actin in the same field stained with TRITC-phalloidin. The unusual bead-like extensions are shown (lower panels). Bars, 10 μm.