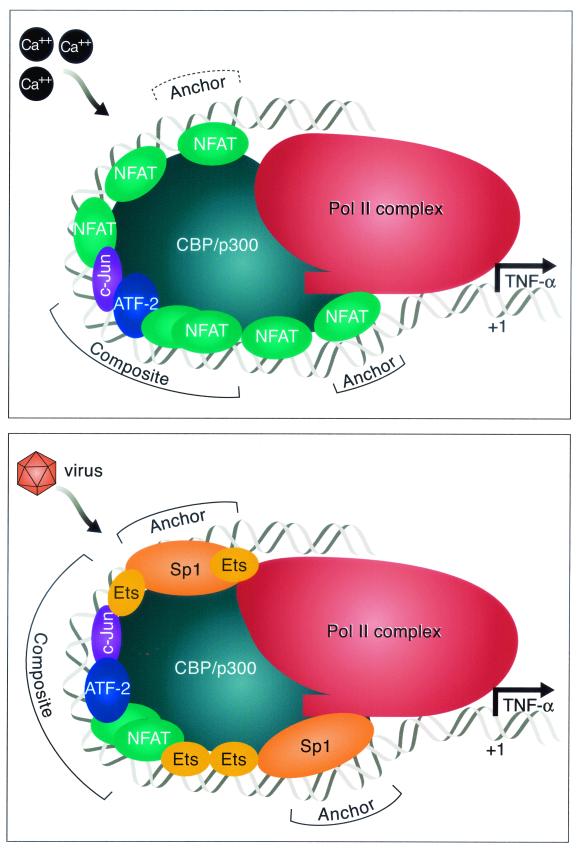
FIG. 7.
Model of the recruitment of inducer-specific TNF-α gene enhanceosomes in T lymphocytes. Distinct enhanceosomes are formed on the TNF-α promoter region in response to ionophore or virus stimulation of T cells. The ionomycin-inducible enhanceosome includes proteins bound to the composite core element CRE/κ3 (ATF-2/c-Jun/NFATp dimer) and two NFAT molecules closest to the basic transcription complex, which we imagine play a major anchoring role in the process of forming the functional transcription-driving complex. We note that phasing mutations upstream of the −76 NFAT site have a relatively modest effect on transcriptional activation by ionomycin, although site-directed mutations in these sites do have a significant impact on gene induction (Fig. 2). The virus-inducible enhanceosome has different components. It has a composite core element ATF-2/c-Jun/NFATp dimer, which also includes Ets protein, and it contains two anchoring Sp1/Ets complexes that, we speculate, are responsible for the recruitment of basic transcription machinery and the activation of transcription. Once assembled, the enhanceosome makes multiple contacts with the basal transcription Pol II complex. The relative sizes of the proteins and the length of the DNA covered are not drawn to scale.