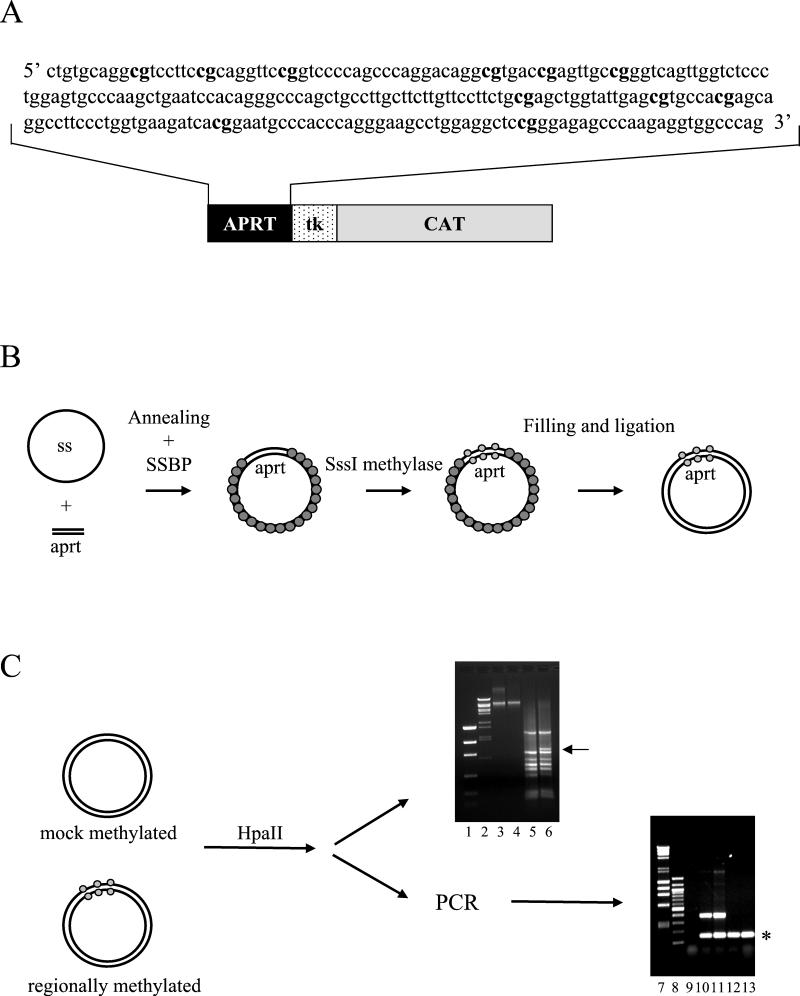
FIG. 1.
Schematic representation of the experimental strategy. (A) The 241-bp sequence amplified from the human aprt CpG island is shown. Methylatable CpG dinucleotides are indicated in bold letters. (B) Scheme of the protocol for the generation of differentially methylated templates. (C) Evaluation of the correct pattern of methylation. The methylation pattern of 11-aprt-tkCAT was analyzed by digestion with the methylation-sensitive restriction enzyme HpaII. The digestion profile was directly visualized on a 1% agarose gel (lanes 5 and 6), or the restricted DNA amplified by PCR (lanes 10 to 13). Markers of low-mass DNA (lane 1), λ BstEII-cut DNA (lanes 2), λ HindIII/φχ HaeIII-cut DNA (lane 7), and 100-bp DNA ladder (lane 8) were resolved. The positions of the supercoiled gel-purified mock-methylated and methylated DNAs are shown in lanes 3 and 4, respectively. Lane 5, HpaII-digested mock-methylated DNA. Lane 6, HpaII-digested regionally methylated DNA. The arrow in lane 6 indicates the appearance of a longer DNA fragment indicative of the incapacity of the enzyme to digest the methylated aprt fragment. PCR amplification of the HpaII-restricted DNAs shown in each lane (10 to 13), and the presence of the internal control is indicated with an asterisk. In contrast, the 450-bp fragment, obtainable only if the aprt fragment is methylated, appears only when the digested regionally methylated DNA is used for the amplification (lanes 10 and 11) and not when mock-methylated DNA is analyzed (lanes 12 and 13). In lane 9, a no-DNA control was loaded.