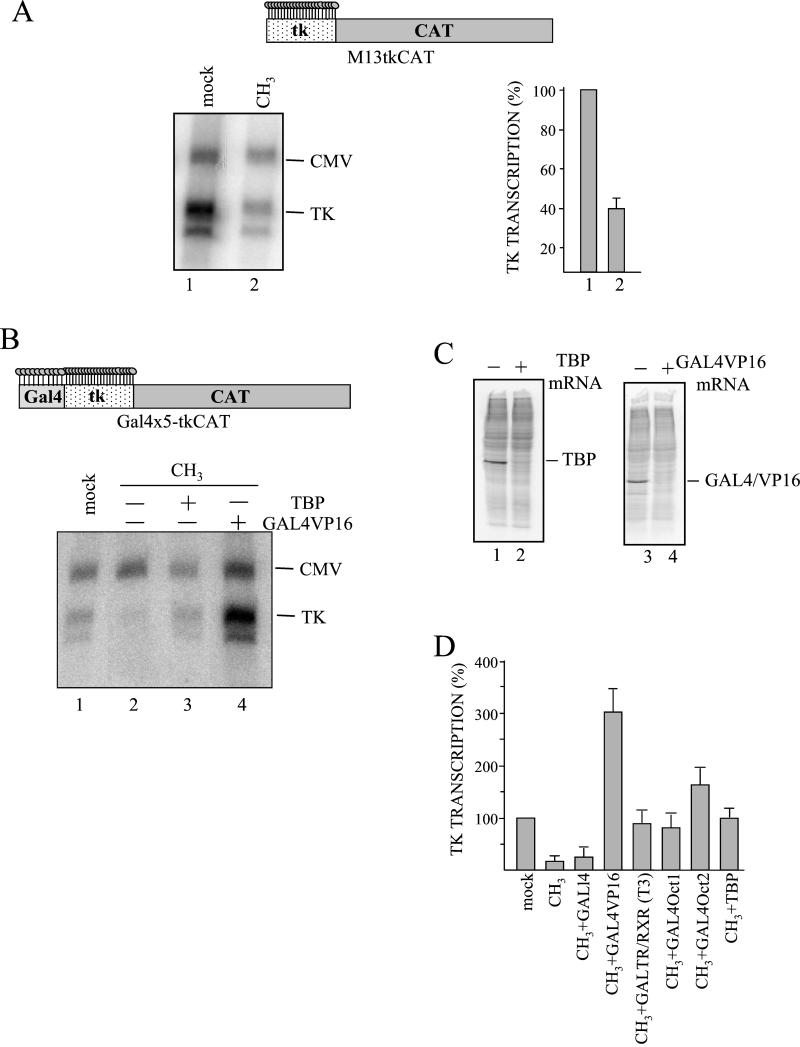
FIG. 6.
Transcriptional activators compete with methylation-specific repressors in vivo. (A) M13-tkCAT vector methylated only on the promoter (lane 2) or the corresponding unmethylated DNA (lane 1) was microinjected. Transcription was evaluated by primer extension and quantitated with a PhosphorImager. Transcriptional levels, relative to the CMV internal standard, were calculated and are reported in the graph as percentages of the 100% active mock DNA. The mean value and standard deviation of three independent experiments are reported. In the upper part of the panel, the regionally methylated M13-tkCAT DNA, characterized by 23 mCpGs distributed over the HSV tk sequence, is represented. (B) TBP (lane 3) or GAL4VP16 (lane 4) mRNA was injected into stage VI oocytes. After incubation for 16 h, a mixture of templates containing unmodified Gal4×5-tkCAT DNA (lane 1) or the same regionally methylated template (lanes 2 to 4) and the CMVCAT internal control was microinjected. Oocytes were collected 16 h after DNA injection, and transcription was analyzed. In the upper part, as above, is a scheme of the partially methylated Gal4×5-tkCAT DNA. (C) A total of 6 ng of TBP (lane 1) or GAL4VP16 (lane 3) mRNAs was injected, and oocytes were incubated for 16 h in the presence of [35S]methionine (100 μCi/ml). Batches of 10 oocytes were collected, and a whole-cell extract (1 oocyte equivalent) from uninjected (lanes 2 and 4) or injected (lanes 1 and 3) oocytes was loaded on SDS-10% PAGE followed by fluorography. The correct positions of exogenously expressed TBP and GAL4VP16 proteins are indicated. (D) A total of 6 ng of a specific mRNA (GAL4, GAL4VP16, a mixture of GALTR and RXR, GAL4OCT1, GAL4OCT2, and TBP) was injected, oocytes were incubated for 16 h and subsequently reinjected with a mixture of regionally methylated Gal4×5-tkCAT and CMV DNAs. A mixture of unmethylated Gal4×5-tkCAT and CMV DNA was also injected as a control (mock). Thyroid hormone (T3, 50 nM) was added to the oocytes synthesizing GALTR/RXR, and all samples were incubated overnight. Transcription was analyzed by primer extension and quantified with a PhosphorImager. The histogram reports the percentage of transcription relative to CMV, calculated from two independent experiments.