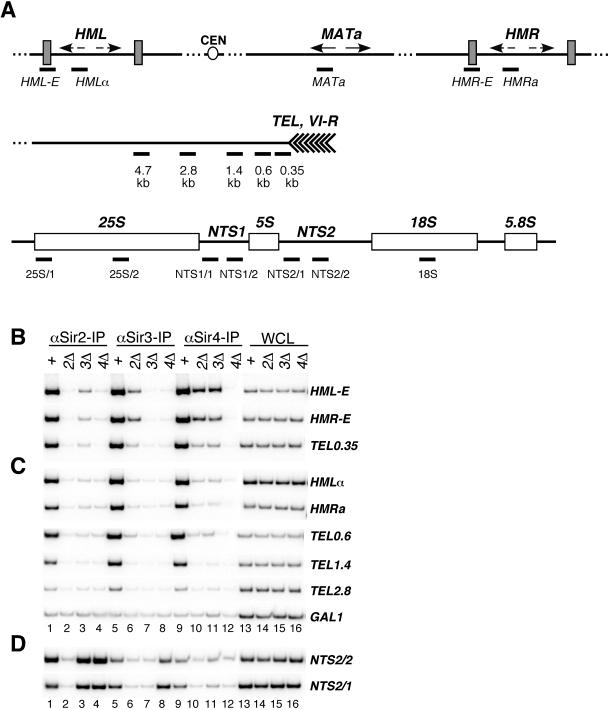
FIG. 2.
Requirement for SIR2, SIR3, and SIR4 for assembly of silent chromatin. (A) Schematic diagram showing the location of PCR primers corresponding to the HML, HMR, and MATa loci on chromosome III, the subtelomeric region on the right arm of chromosome VI (TEL and VI-R), and the rDNA repeats on chromosome XII used in chromatin immunoprecipitation experiments. The locations of PCR primers are indicated under each region as thick bars. The telomeric primers amplify DNA fragments from 0.35, 0.6, 1.4, 2.8, and 4.7 kb from the telomeric repeats. The HML and HMR primers flank the E silencers (HML-E and HMR-E) or are located within HMLα or HMRa, respectively. The rDNA primers amplify fragments within the nontranscribed spacer regions (NTS1 and NTS2) and the 35S rRNA coding region. Strains used were the wild type (W303-1a) and sir2Δ (SF3), sir3Δ (SF4), and sir4Δ (SF5) mutants. (B) Phosphorimager data of chromatin immunoprecipitation experiments showing the association of the Sir2, Sir3, and Sir4 proteins with silencers (HML-E and HMR-E) and DNA immediately adjacent to telomere VI-R in wild-type (lanes +), sir2Δ (2Δ), sir3Δ (3Δ), and sir4Δ (4Δ) strains. PCR amplifications of anti-Sir2, anti-Sir3, and anti-Sir4 chromatin immunoprecipitations and input DNA from WCL are shown. (C) Association of Sir2, Sir3, and Sir4 in the same genetic backgrounds as in B with silent chromatin regions that are more distal from initiation sites and with a control nonsilenced locus (GAL1). (D) Association of Sir2, Sir3, and Sir4 with rDNA.