FIG. 4.
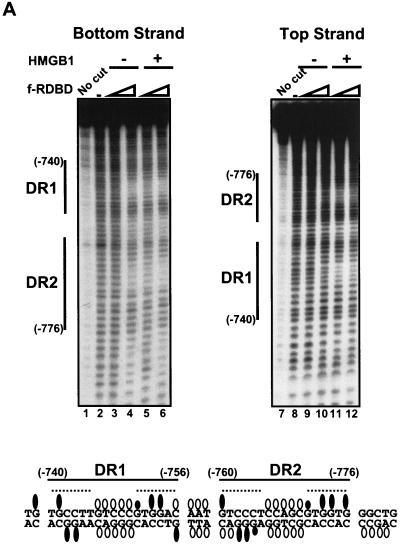
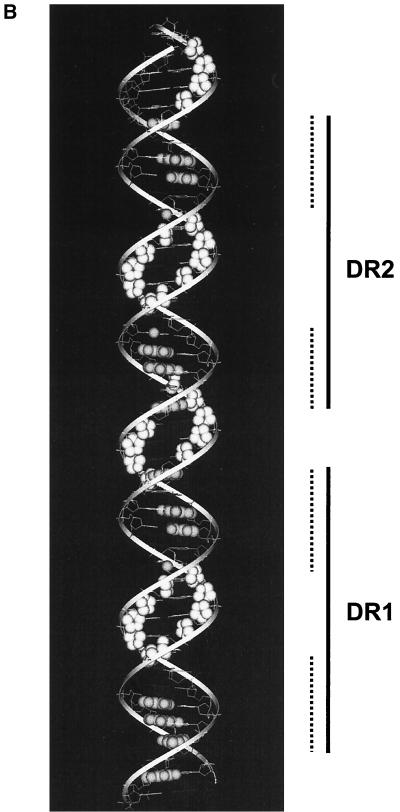
HMGB1 does not form a stable ternary complex with f-RDBD and DNA. (A) Hydroxyl radical footprinting reveals the interactions of f-RDBD with the minor groove, but no HMGB1 specific interactions; 7 fmol of an 85-bp fragment containing DR1 and DR2 was 32P end labeled on the bottom (left panel) or the top strand (right panel) and incubated in the presence of increasing amounts of f-RDBD (9.7 ng or 13.5 nM, lanes 3, 5, 9, and 11; 132 ng or 185 nM, lanes 4, 6, 10, and 12) and 7.7 μg of poly(dI-dC) per ml. Then 712 ng (1.8 μM) of HMGB1 was added to lanes 5 to 6 and 11 to 12. The positions of the sites relative to the start site of transcription are also shown. The bands protected by f-RDBD from hydroxyl radical cleavage were quantitated and identified as described in Materials and Methods. DNase I footprinting reactions were performed in parallel under the hydroxyl radical binding conditions to ensure f-RDBD binding and that the helping effect of HMGB1 was observed (data not shown). The bottom panel summarizes the hydroxyl radical protections (open circles) identified above. Guanines in DR1 and DR2 that interfere with Rta binding in methylation experiments by Gruffat and Sergeant (24) are shown as black ovals. The size of the ovals corresponds to the degree of interference. The dashed lines indicate the DR1 and DR2 half-sites, which are presumably contacted by Rta monomers within the dimer (24). (B) Modeling of the hydroxyl radical and DMS interference results summarized in the previous panel. A helical projection of the region of the BHLF-1 enhancer encompassing DR1 and DR2 was generated using InsightII. The sugars attacked by hydroxyl radical cleavage are highlighted in white. Guanines shown to interfere with Rta binding by Gruffat and Sergeant (24) are highlighted in gray. Guanines that interfere to a lesser extent are only highlighted on N7. The dashed lines indicate the DR1 and DR2 half sites.