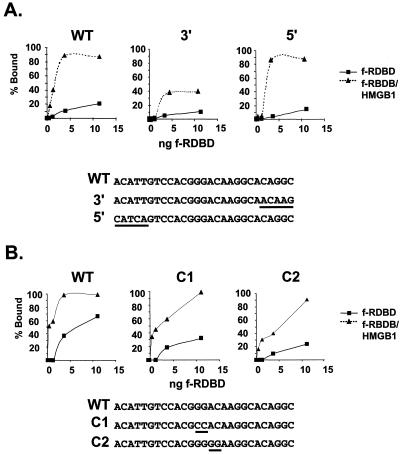
FIG. 6.
Sequence requirements for the stimulatory effect of HMGB1. (A) Mutations in the 5′ and 3′ DR1 flanking sequences do not abolish the stimulatory effect of HMGB1; 1 fmol of radiolabeled wild-type (WT) or 5′ or 3′ mutant oligonucleotides was incubated in the presence of increasing concentrations of f-RDBD (0.4, 1.2, 3.6, and 11.1 ng or 0.6, 1.7, 5, and 15 nM) and 7.7 μg of poly(dI-dC) per ml, in the presence or absence of 158 ng (0.4 μM) of recombinant HMGB1. The reaction mixtures were then subjected to EMSA. The percentages of probe bound by f-RDBD in the absence (rectangles) and presence (triangles) of HMGB1 were calculated and plotted against the amount of f-RDBD used. The graphs represent data points from the same gel. The sequences of the oligonucleotides are shown below the graphs. The 5-bp mutations are underlined. (B) Mutations in the central, nonconserved region of DR1 do not affect HMGB1-mediated cooperativity. Reactions were performed as described above using the WT or the C1 or C2 mutant oligonucleotide. The reactions were quantitated and plotted as described above. The sequences of the oligonucleotides are shown below the graphs. The 2-bp mutations are underlined.