FIG.8.
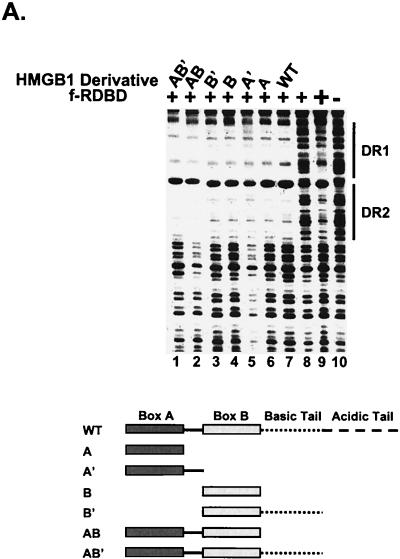
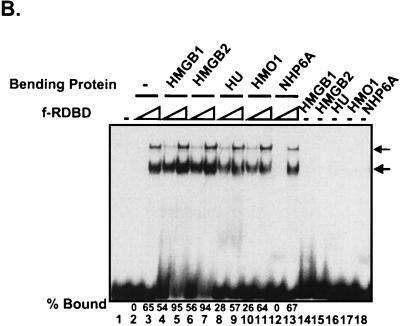
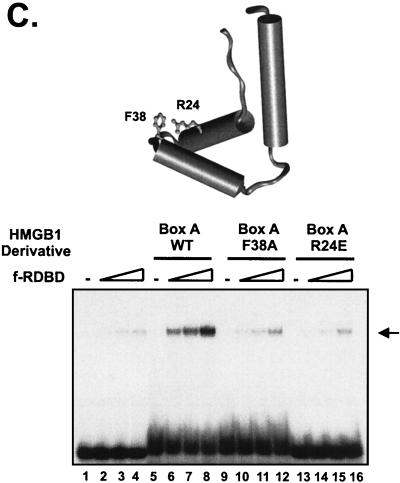
Variety of DNA-bending proteins can facilitate Rta binding. (A) Deletion of the individual DNA-binding domains of HMGB1 has no effect on the stimulation of f-RDBD binding; 5 fmol of a 110-bp radiolabeled restriction fragment containing the DR1 and DR2 binding sites was incubated in the presence of saturating (60 ng or 84 nM, lane 9) and subsaturating (15 ng or 21 nM, lanes 1 to 8) amounts of f-RDBD and 7.7 μg of poly(dI-dC) per ml. The following deletion derivatives of HMGB1 were added: 316 ng (0.84 μM, lane 7) of wild-type HMGB1, 333 ng (lane 6) of A box, 333 ng (lane 5) of A′ box, 712 ng (lane 4) of B box, 333 ng (lane 3) of B′ box, 712 ng (lane 2) of AB box, and 712 ng (lane 1) of AB′ box. The schematic in the bottom panel shows the domain organization of the various deletion derivatives. Each deletion mutant was initially titrated with f-RDBD, and the amount eliciting the greatest response was chosen for subsequent experiments. (B) Eukaryotic and prokaryotic DNA-bending proteins promote f-RDBD binding to DR1; 1 fmol of the wild type DR1 oligonucleotide was incubated in the presence of increasing amounts of f-RDBD (0.4 and 11.1 ng or 0.56 and 15 nM, respectively; lanes 2 and 3, 4 and 5, 6 and 7, 8 and 9, 10 and 11, and 12 and 13) and 7.7 μg of poly(dI-dC) per ml. The following DNA-bending proteins were added: 158 ng of HMGB1 (0.4 μM, lanes 4, 5, and 14), 333 ng of HMGB2 (0.85 μM, lanes 6, 7, and 15), 110 ng of HU (0.47 μM, lanes 8, 9, and 16), 53 ng of HMO1 (0.16 μM, lanes 10, 11, and 17), and 21 ng of NHP6A (0.15 μM, lanes 12, 13, and 18). The percent probe bound by f-RDBD was calculated as described for Fig. 3C. The bending proteins were initially titrated with f-RDBD, and the concentrations that elicited the maximal helping effect were chosen for subsequent experiments. (C) Mutations in the HMGB1 A box that affect DNA bending and DNA binding abolish the helping effect on f-RDBD binding. The schematic illustration of the box A structure (44) indicates the position of the three α-helices in box A and the positions of phenylalanine 38, which was mutated to an alanine, and arginine 24, which was replaced with a glutamic acid. The proteins were expressed and purified as described in Materials and Methods. The wild-type and mutant proteins were assayed by EMSA for their ability to facilitate f-RDBD binding (bottom panel); 1 fmol of radiolabeled DR1 oligonucleotide was incubated with increasing amounts of f-RDBD (0.2, 0.4, and 1.2 ng or 0.3, 0.6, and 1.7 nM) and 7.7 μg of poly(dI-dC) per ml. Then 150 ng (0.18 μM) of box A wild-type or mutants was added to the reaction mixtures as indicated. Note that addition of the DNA-bending mutant (F38A) causes a slight upward smear of the probe in a manner similar to wild-type box A, which reflects their ability to form unstable complexes with DNA. The DNA-binding mutant R24E does not smear the probe.